Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL0222 [new locus tag: SACOL_RS01135 ]
- pan locus tag?: SAUPAN001107000
- symbol: ldh1
- pan gene symbol?: ldh1
- synonym:
- product: L-lactate dehydrogenase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL0222 [new locus tag: SACOL_RS01135 ]
- symbol: ldh1
- product: L-lactate dehydrogenase
- replicon: chromosome
- strand: +
- coordinates: 262250..263203
- length: 954
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3236813 NCBI
- RefSeq: YP_185120 NCBI
- BioCyc: see SACOL_RS01135
- MicrobesOnline: 911698 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901ATGAACAAATTTAAAGGGAACAAAGTTGTATTAATAGGTAATGGTGCAGTAGGTTCAAGC
TACGCATTTTCATTAGTGAACCAAAGCATTGTTGATGAATTAGTCATCATTGATTTAGAC
ACTGAAAAAGTTCGAGGAGATGTTATGGATTTAAAACATGCCACACCATATTCTCCAACA
ACAGTTCGTGTGAAAGCTGGCGAATACAGTGATTGTCATGATGCGGATCTAGTTGTCATC
TGTGCTGGTGCTGCACAAAAACCTGGAGAAACACGTTTAGATTTAGTATCTAAAAACTTG
AAAATATTCAAATCAATTGTTGGTGAAGTAATGGCATCAAAATTTGATGGTATTTTCTTG
GTAGCTACAAATCCTGTTGATATTTTAGCGTATGCAACATGGAAATTCTCTGGTTTACCT
AAAGAACGTGTTATAGGTTCTGGTACAATTTTAGACTCTGCACGCTTTAGATTATTGTTA
AGCGAAGCGTTCGATGTTGCGCCACGTAGCGTCGATGCTCAAATTATTGGTGAACATGGT
GACACTGAATTACCAGTATGGTCACACGCTAATATTGCGGGTCAACCTTTGAAGACATTA
CTTGAACAACGTCCTGAGGGCAAAGCGCAAATTGAACAAATTTTTGTTCAAACACGTGAT
GCAGCATATGACATTATTCAAGCTAAAGGTGCCACTTATTATGGTGTTGCAATGGGATTA
GCTAGAATTACTGAAGCGATTTTCAGAAATGAAGATGCCGTATTGACTGTATCAGCATTA
TTAGAAGGCGAATATGAGGAAGAAGATGTTTATATTGGTGTTCCAGCAGTCATCAATAGA
AACGGTATTCGCAACGTCGTAGAAATCCCATTAAACGACGAAGAACAAAGCAAGTTCGCA
CATTCAGCTAAAACATTAAAAGATATTATGGCTGAAGCAGAAGAACTTAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
954
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL0222 [new locus tag: SACOL_RS01135 ]
- symbol: Ldh1
- description: L-lactate dehydrogenase
- length: 317
- theoretical pI: 4.6926
- theoretical MW: 34583.2
- GRAVY: -0.0312303
⊟Function[edit | edit source]
- reaction: EC 1.1.1.27? ExPASyL-lactate dehydrogenase (S)-lactate + NAD+ = pyruvate + NADH
- TIGRFAM: Energy metabolism Anaerobic L-lactate dehydrogenase (TIGR01771; EC 1.1.1.27; HMM-score: 433.4)Energy metabolism Glycolysis/gluconeogenesis L-lactate dehydrogenase (TIGR01771; EC 1.1.1.27; HMM-score: 433.4)and 9 moreEnergy metabolism TCA cycle malate dehydrogenase, NAD-dependent (TIGR01763; EC 1.1.1.37; HMM-score: 239.8)Energy metabolism TCA cycle malate dehydrogenase, NAD-dependent (TIGR01772; EC 1.1.1.37; HMM-score: 78.7)malate dehydrogenase (TIGR01759; EC 1.1.1.-; HMM-score: 68)malate dehydrogenase, NAD-dependent (TIGR01758; EC 1.1.1.37; HMM-score: 53.5)lactate dehydrogenase (TIGR01756; EC 1.1.1.27; HMM-score: 38)malate dehydrogenase, NADP-dependent (TIGR01757; EC 1.1.1.82; HMM-score: 21.5)Protein fate Protein modification and repair coenzyme F420-reducing hydrogenase, FrhD protein (TIGR00130; HMM-score: 14.9)Biosynthesis of cofactors, prosthetic groups, and carriers Thiamine thiazole biosynthesis adenylyltransferase ThiF (TIGR02356; EC 2.7.7.73; HMM-score: 13.6)Biosynthesis of cofactors, prosthetic groups, and carriers Pantothenate and coenzyme A 2-dehydropantoate 2-reductase (TIGR00745; EC 1.1.1.-; HMM-score: 11.8)
- TheSEED :
- Malate dehydrogenase (EC 1.1.1.37)
and 1 more - PFAM: NADP_Rossmann (CL0063) Ldh_1_N; lactate/malate dehydrogenase, NAD binding domain (PF00056; HMM-score: 169.6)LDH_C (CL0341) Ldh_1_C; lactate/malate dehydrogenase, alpha/beta C-terminal domain (PF02866; HMM-score: 166.6)and 4 moreNADP_Rossmann (CL0063) NAD_binding_7; Putative NAD(P)-binding (PF13241; HMM-score: 15.9)Semialdhyde_dh; Semialdehyde dehydrogenase, NAD binding domain (PF01118; HMM-score: 14.1)Shikimate_DH; Shikimate / quinate 5-dehydrogenase (PF01488; HMM-score: 14)Sacchrp_dh_NADP; Saccharopine dehydrogenase NADP binding domain (PF03435; HMM-score: 13.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9764
- Cytoplasmic Membrane Score: 0.0055
- Cell wall & surface Score: 0.0008
- Extracellular Score: 0.0173
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.149003
- TAT(Tat/SPI): 0.001671
- LIPO(Sec/SPII): 0.01365
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNKFKGNKVVLIGNGAVGSSYAFSLVNQSIVDELVIIDLDTEKVRGDVMDLKHATPYSPTTVRVKAGEYSDCHDADLVVICAGAAQKPGETRLDLVSKNLKIFKSIVGEVMASKFDGIFLVATNPVDILAYATWKFSGLPKERVIGSGTILDSARFRLLLSEAFDVAPRSVDAQIIGEHGDTELPVWSHANIAGQPLKTLLEQRPEGKAQIEQIFVQTRDAAYDIIQAKGATYYGVAMGLARITEAIFRNEDAVLTVSALLEGEYEEEDVYIGVPAVINRNGIRNVVEIPLNDEEQSKFAHSAKTLKDIMAEAEELK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas
- protein localization: Cytoplasmic [1] [2] [3]
- quantitative data / protein copy number per cell: 46 [4]
- interaction partners:
SACOL1760 (ackA) acetate kinase [5] (data from MRSA252) SACOL0660 (adhP) alcohol dehydrogenase [5] (data from MRSA252) SACOL0452 (ahpC) alkyl hydroperoxide reductase subunit C [5] (data from MRSA252) SACOL2657 (arcA) arginine deiminase [5] (data from MRSA252) SACOL2656 (arcB2) ornithine carbamoyltransferase [5] (data from MRSA252) SACOL2654 (arcC2) carbamate kinase [5] (data from MRSA252) SACOL0557 (cysK) cysteine synthase [5] (data from MRSA252) SACOL1800 (dat) D-alanine aminotransferase [5] (data from MRSA252) SACOL0123 (deoC1) deoxyribose-phosphate aldolase [5] (data from MRSA252) SACOL1399 (dmpI) 4-oxalocrotonate tautomerase [5] (data from MRSA252) SACOL1637 (dnaK) molecular chaperone DnaK [5] (data from MRSA252) SACOL0842 (eno) phosphopyruvate hydratase [5] (data from MRSA252) SACOL0634 (eutD) phosphotransacetylase [5] (data from MRSA252) SACOL0988 (fabF) 3-oxoacyl-ACP synthase [5] (data from MRSA252) SACOL1016 (fabI) enoyl-ACP reductase [5] (data from MRSA252) SACOL2117 (fbaA) fructose-bisphosphate aldolase [5] (data from MRSA252) SACOL2622 (fdaB) fructose-1,6-bisphosphate aldolase [5] (data from MRSA252) SACOL1329 (femC) glutamine synthetase [5] (data from MRSA252) SACOL1782 (fhs) formate--tetrahydrofolate ligase [5] (data from MRSA252) SACOL1199 (ftsZ) cell division protein FtsZ [5] (data from MRSA252) SACOL0593 (fusA) elongation factor G [5] (data from MRSA252) SACOL0838 (gapA1) glyceraldehyde 3-phosphate dehydrogenase [5] (data from MRSA252) SACOL1960 (gatB) aspartyl/glutamyl-tRNA amidotransferase subunit B [5] (data from MRSA252) SACOL0877 (gcvH) glycine cleavage system protein H [5] (data from MRSA252) SACOL2145 (glmS) glucosamine--fructose-6-phosphate aminotransferase [5] (data from MRSA252) SACOL1742 (gltA) citrate synthase [5] (data from MRSA252) SACOL0961 (gluD) glutamate dehydrogenase [5] (data from MRSA252) SACOL1554 (gnd) 6-phosphogluconate dehydrogenase [5] (data from MRSA252) SACOL2415 (gpmA) phosphoglyceromutase [5] (data from MRSA252) SACOL1638 (grpE) heat shock protein GrpE [5] (data from MRSA252) SACOL0461 (guaA) GMP synthase [5] (data from MRSA252) SACOL0460 (guaB) inosine-5'-monophosphate dehydrogenase [5] (data from MRSA252) SACOL1513 (hup) DNA-binding protein HU [5] (data from MRSA252) SACOL1741 (icd) isocitrate dehydrogenase [5] (data from MRSA252) SACOL1206 (ileS) isoleucyl-tRNA synthetase [5] (data from MRSA252) SACOL1477 (ilvA1) threonine dehydratase [5] (data from MRSA252) SACOL1288 (infB) translation initiation factor IF-2 [5] (data from MRSA252) SACOL1368 (kataA) catalase [5] (data from MRSA252) SACOL2618 (ldh2) L-lactate dehydrogenase [5] (data from MRSA252) SACOL2623 (mqo2) malate:quinone oxidoreductase [5] (data from MRSA252) SACOL2116 (murAB) UDP-N-acetylglucosamine 1-carboxyvinyltransferase [5] (data from MRSA252) SACOL0746 (norR) MarR family transcriptional regulator [5] (data from MRSA252) SACOL1102 (pdhA) pyruvate dehydrogenase complex E1 component subunit alpha [5] (data from MRSA252) SACOL1103 (pdhB) pyruvate dehydrogenase complex E1 component subunit beta [5] (data from MRSA252) SACOL1104 (pdhC) branched-chain alpha-keto acid dehydrogenase E2 [5] (data from MRSA252) SACOL1105 (pdhD) dihydrolipoamide dehydrogenase [5] (data from MRSA252) SACOL2128 (pdp) pyrimidine-nucleoside phosphorylase [5] (data from MRSA252) SACOL1746 (pfkA) 6-phosphofructokinase [5] (data from MRSA252) SACOL0204 (pflB) formate acetyltransferase [5] (data from MRSA252) SACOL0966 (pgi) glucose-6-phosphate isomerase [5] (data from MRSA252) SACOL0839 (pgk) phosphoglycerate kinase [5] (data from MRSA252) SACOL0841 (pgm) phosphoglyceromutase [5] (data from MRSA252) SACOL1293 (pnp) polynucleotide phosphorylase/polyadenylase [5] (data from MRSA252) SACOL1282 (proS) prolyl-tRNA synthetase [5] (data from MRSA252) SACOL1745 (pyk) pyruvate kinase [5] (data from MRSA252) SACOL2119 (pyrG) CTP synthetase [5] (data from MRSA252) SACOL0584 (rplA) 50S ribosomal protein L1 [5] (data from MRSA252) SACOL2236 (rplB) 50S ribosomal protein L2 [5] (data from MRSA252) SACOL2227 (rplE) 50S ribosomal protein L5 [5] (data from MRSA252) SACOL0585 (rplJ) 50S ribosomal protein L10 [5] (data from MRSA252) SACOL0583 (rplK) 50S ribosomal protein L11 [5] (data from MRSA252) SACOL2207 (rplM) 50S ribosomal protein L13 [5] (data from MRSA252) SACOL2220 (rplO) 50S ribosomal protein L15 [5] (data from MRSA252) SACOL2212 (rplQ) 50S ribosomal protein L17 [5] (data from MRSA252) SACOL1257 (rplS) 50S ribosomal protein L19 [5] (data from MRSA252) SACOL1725 (rplT) 50S ribosomal protein L20 [5] (data from MRSA252) SACOL1702 (rplU) 50S ribosomal protein L21 [5] (data from MRSA252) SACOL2234 (rplV) 50S ribosomal protein L22 [5] (data from MRSA252) SACOL2228 (rplX) 50S ribosomal protein L24 [5] (data from MRSA252) SACOL0589 (rpoC) DNA-directed RNA polymerase subunit beta' [5] (data from MRSA252) SACOL2120 (rpoE) DNA-directed RNA polymerase subunit delta [5] (data from MRSA252) SACOL1274 (rpsB) 30S ribosomal protein S2 [5] (data from MRSA252) SACOL2233 (rpsC) 30S ribosomal protein S3 [5] (data from MRSA252) SACOL1769 (rpsD) 30S ribosomal protein S4 [5] (data from MRSA252) SACOL2222 (rpsE) 30S ribosomal protein S5 [5] (data from MRSA252) SACOL0592 (rpsG) 30S ribosomal protein S7 [5] (data from MRSA252) SACOL2225 (rpsH) 30S ribosomal protein S8 [5] (data from MRSA252) SACOL2206 (rpsI) 30S ribosomal protein S9 [5] (data from MRSA252) SACOL2214 (rpsK) 30S ribosomal protein S11 [5] (data from MRSA252) SACOL0095 (spa) immunoglobulin G binding protein A precursor [5] (data from MRSA252) SACOL0541 (spoVG) regulatory protein SpoVG [5] (data from MRSA252) SACOL1448 (sucB) dihydrolipoamide succinyltransferase [5] (data from MRSA252) SACOL1262 (sucC) succinyl-CoA synthetase subunit beta [5] (data from MRSA252) SACOL1263 (sucD) succinyl-CoA synthetase subunit alpha [5] (data from MRSA252) SACOL1831 (tal) translaldolase [5] (data from MRSA252) SACOL0626 (thiD1) phosphomethylpyrimidine kinase [5] (data from MRSA252) SACOL1722 (tig) trigger factor [5] (data from MRSA252) SACOL1377 (tkt) transketolase [5] (data from MRSA252) SACOL1762 (tpx) thiol peroxidase [5] (data from MRSA252) SACOL1155 (trxA) thioredoxin [5] (data from MRSA252) SACOL1276 (tsf) elongation factor Ts [5] (data from MRSA252) SACOL0594 (tuf) elongation factor Tu [5] (data from MRSA252) SACOL1118 (typA) GTP-binding protein TypA [5] (data from MRSA252) SACOL2104 (upp) uracil phosphoribosyltransferase [5] (data from MRSA252) SACOL0303 5'-nucleotidase [5] (data from MRSA252) SACOL0521 hypothetical protein [5] (data from MRSA252) SACOL0565 glutamine amidotransferase subunit PdxT [5] (data from MRSA252) SACOL0688 ABC transporter substrate-binding protein [5] (data from MRSA252) SACOL0708 DAK2 domain-containing protein [5] (data from MRSA252) SACOL0721 hypothetical protein [5] (data from MRSA252) SACOL0731 LysR family transcriptional regulator [5] (data from MRSA252) SACOL0815 ribosomal subunit interface protein [5] (data from MRSA252) SACOL0875 thioredoxin [5] (data from MRSA252) SACOL0944 NADH dehydrogenase [5] (data from MRSA252) SACOL0973 fumarylacetoacetate hydrolase [5] (data from MRSA252) SACOL1099 hypothetical protein [5] (data from MRSA252) SACOL1437 CSD family cold shock protein [5] (data from MRSA252) SACOL1457 PTS system, IIA component [5] (data from MRSA252) SACOL1753 universal stress protein [5] (data from MRSA252) SACOL1759 universal stress protein [5] (data from MRSA252) SACOL1801 dipeptidase PepV [5] (data from MRSA252) SACOL1891 RNAIII-activating protein TRAP [5] (data from MRSA252) SACOL1952 ferritins family protein [5] (data from MRSA252) SACOL2173 alkaline shock protein 23 [5] (data from MRSA252) SACOL2296 glycerate dehydrogenase [5] (data from MRSA252) SACOL2535 D-lactate dehydrogenase [5] (data from MRSA252) SACOL2569 1-pyrroline-5-carboxylate dehydrogenase [5] (data from MRSA252)
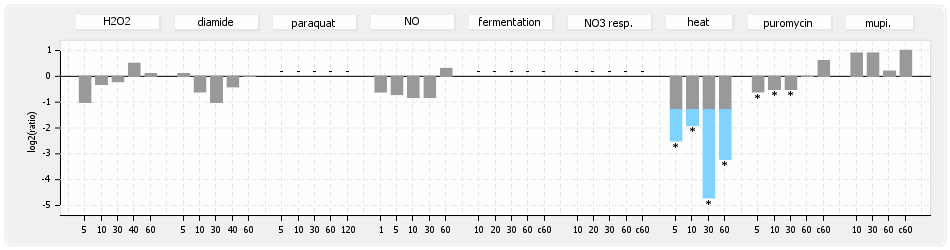
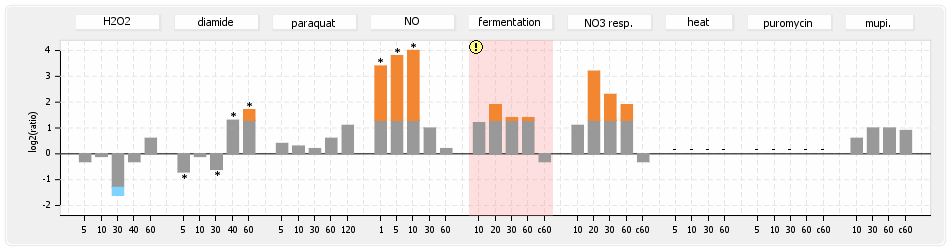
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator: Rex* (repression) regulon
Rex* (TF) important in Energy metabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: data available for NCTC8325
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: 53.15 h [6]
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ 5.000 5.001 5.002 5.003 5.004 5.005 5.006 5.007 5.008 5.009 5.010 5.011 5.012 5.013 5.014 5.015 5.016 5.017 5.018 5.019 5.020 5.021 5.022 5.023 5.024 5.025 5.026 5.027 5.028 5.029 5.030 5.031 5.032 5.033 5.034 5.035 5.036 5.037 5.038 5.039 5.040 5.041 5.042 5.043 5.044 5.045 5.046 5.047 5.048 5.049 5.050 5.051 5.052 5.053 5.054 5.055 5.056 5.057 5.058 5.059 5.060 5.061 5.062 5.063 5.064 5.065 5.066 5.067 5.068 5.069 5.070 5.071 5.072 5.073 5.074 5.075 5.076 5.077 5.078 5.079 5.080 5.081 5.082 5.083 5.084 5.085 5.086 5.087 5.088 5.089 5.090 5.091 5.092 5.093 5.094 5.095 5.096 5.097 5.098 5.099 5.100 5.101 5.102 5.103 5.104 5.105 5.106 5.107 5.108 5.109 5.110 5.111 5.112 5.113 5.114 5.115 5.116 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Jörg Bernhardt, Andreas Otto, Martin Moche, Dörte Becher, Hanna Meyer, Michael Lalk, Claudia Schurmann, Rabea Schlüter, Holger Kock, Ulf Gerth, Michael Hecker
Life and death of proteins: a case study of glucose-starved Staphylococcus aureus.
Mol Cell Proteomics: 2012, 11(9);558-70
[PubMed:22556279] [WorldCat.org] [DOI] (I p)
⊟Relevant publications[edit | edit source]
Anthony R Richardson, Stephen J Libby, Ferric C Fang
A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity.
Science: 2008, 319(5870);1672-6
[PubMed:18356528] [WorldCat.org] [DOI] (I p)Jia Bin Dong, Xiang Liu, Lan Fen Li, Shangwei Wu, Xiao Dong Su
Preliminary X-ray crystallographic analysis of a nitric oxide-inducible lactate dehydrogenase from Staphylococcus aureus.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2009, 65(Pt 10);1053-5
[PubMed:19851020] [WorldCat.org] [DOI] (I p)