⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01391
- pan locus tag?: SAUPAN003803000
- symbol: SAOUHSC_01391
- pan gene symbol?: cvfB
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01391
- symbol: SAOUHSC_01391
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1333318..1334220
- length: 903
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920682 NCBI
- RefSeq: YP_499919 NCBI
- BioCyc: G1I0R-1300 BioCyc
- MicrobesOnline: 1289833 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901ATGGCATTAGACAAAGATATAGTAGGTTCTATAGAATTCCTTGAAGTAGTAGGGTTACAA
GGTTCAACTTACCTTTTAAAAGGACCAAACGGTGAAAACGTAAAGTTAAACCAATCAGAA
ATGAACGATGATGATGAATTAGAAGTAGGTGAAGAATATAGTTTCTTCATTTATCCAAAC
CGTTCAGGTGAATTATTTGCAACTCAAAATATGCCTGATATTACGAAAGATAAATATGAC
TTTGCTAAAGTACTTAAAACGGATCGCGATGGGGCACGTATAGATGTTGGATTACCCCGT
GAAGTGTTAGTACCATGGGAAGATTTACCAAAAGTGAAATCACTATGGCCACAACCTGGT
GATTATTTGCTAGTTACATTACGAATTGACCGTGAGAATCATATGTATGGACGTTTAGCG
AGTGAATCTGTTGTAGAAAATATGTTTACACCTGTACACGACGATAATTTAAAAAACGAA
GTCATTGAAGCCAAACCTTACCGCGTATTACGAATTGGTAGCTTTTTATTAAGCGAATCA
GGTTACAAAATTTTCGTACATGAATCAGAACGTAAAGCTGAACCAAGATTAGGTGAATCT
GTTCAAGTTAGAATTATCGGGCATAATGATAAAGGTGAGTTAAATGGTTCATTTTTACCA
CTTGCACATGAACGTTTAGACGATGACGGCCAAGTCATCTTTGATTTACTAGTTGAATAT
GATGGTGAATTACCATTCTGGGACAAATCAAGCCCTGAAGCGATTAAAGAAGTATTCAAT
ATGAGTAAAGGTTCATTCAAACGTGCAATCGGTCACTTATATAAACAGAAGATTATTAAT
ATAGAAACAGGTAAAATCGCTTTAACTAAAAAAGGTTGGAGTCGAATGGACTCAAAAGAA
TAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
903
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01391
- symbol: SAOUHSC_01391
- description: hypothetical protein
- length: 300
- theoretical pI: 4.76461
- theoretical MW: 34198.5
- GRAVY: -0.494667
⊟Function[edit | edit source]
- TIGRFAM:
- TheSEED :
- S1 RNA binding domain
- PFAM: HTH (CL0123) CvfB_WH; CvfB-like winged helix domain (PF17783; HMM-score: 78.8)and 4 moreOB (CL0021) CvfB_2nd; Virulence regulatory factor CvfB, second domain (PF21543; HMM-score: 62.2)CvfB_1st; CvfB first S1-like domain (PF21191; HMM-score: 52)S1_2; S1 domain (PF13509; HMM-score: 39.3)S1; S1 RNA binding domain (PF00575; HMM-score: 17.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9778
- Cytoplasmic Membrane Score: 0.0167
- Cell wall & surface Score: 0.0002
- Extracellular Score: 0.0053
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.005867
- TAT(Tat/SPI): 0.002812
- LIPO(Sec/SPII): 0.002347
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MALDKDIVGSIEFLEVVGLQGSTYLLKGPNGENVKLNQSEMNDDDELEVGEEYSFFIYPNRSGELFATQNMPDITKDKYDFAKVLKTDRDGARIDVGLPREVLVPWEDLPKVKSLWPQPGDYLLVTLRIDRENHMYGRLASESVVENMFTPVHDDNLKNEVIEAKPYRVLRIGSFLLSESGYKIFVHESERKAEPRLGESVQVRIIGHNDKGELNGSFLPLAHERLDDDGQVIFDLLVEYDGELPFWDKSSPEAIKEVFNMSKGSFKRAIGHLYKQKIINIETGKIALTKKGWSRMDSKE
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_01201 (acpP) acyl carrier protein [3] (data from MRSA252) SAOUHSC_02490 (adk) adenylate kinase [3] (data from MRSA252) SAOUHSC_00611 (argS) arginyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01170 (carB) carbamoyl phosphate synthase large subunit [3] (data from MRSA252) SAOUHSC_01496 (cmk) cytidylate kinase [3] (data from MRSA252) SAOUHSC_02318 (ddl) D-alanyl-alanine synthetase A [3] (data from MRSA252) SAOUHSC_02336 (fabZ) (3R)-hydroxymyristoyl-ACP dehydratase [3] (data from MRSA252) SAOUHSC_01236 (frr) ribosome recycling factor [3] (data from MRSA252) SAOUHSC_02117 (gatA) aspartyl/glutamyl-tRNA amidotransferase subunit A [3] (data from MRSA252) SAOUHSC_02116 (gatB) aspartyl/glutamyl-tRNA amidotransferase subunit B [3] (data from MRSA252) SAOUHSC_01634 (gcvT) glycine cleavage system aminomethyltransferase T [3] (data from MRSA252) SAOUHSC_02255 (groES) co-chaperonin GroES [3] (data from MRSA252) SAOUHSC_01159 (ileS) isoleucyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01246 (infB) translation initiation factor IF-2 [3] (data from MRSA252) SAOUHSC_01786 (infC) translation initiation factor IF-3 [3] (data from MRSA252) SAOUHSC_01875 (leuS) leucyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01485 (ndk) nucleoside diphosphate kinase [3] (data from MRSA252) SAOUHSC_01243 (nusA) transcription elongation factor NusA [3] (data from MRSA252) SAOUHSC_02919 (panB) 3-methyl-2-oxobutanoate hydroxymethyltransferase [3] (data from MRSA252) SAOUHSC_01168 (pyrC) dihydroorotase [3] (data from MRSA252) SAOUHSC_00519 (rplA) 50S ribosomal protein L1 [3] (data from MRSA252) SAOUHSC_02509 (rplB) 50S ribosomal protein L2 [3] (data from MRSA252) SAOUHSC_02512 (rplC) 50S ribosomal protein L3 [3] (data from MRSA252) SAOUHSC_02511 (rplD) 50S ribosomal protein L4 [3] (data from MRSA252) SAOUHSC_02500 (rplE) 50S ribosomal protein L5 [3] (data from MRSA252) SAOUHSC_02496 (rplF) 50S ribosomal protein L6 [3] (data from MRSA252) SAOUHSC_00017 (rplI) 50S ribosomal protein L9 [3] (data from MRSA252) SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [3] (data from MRSA252) SAOUHSC_00521 (rplL) 50S ribosomal protein L7/L12 [3] (data from MRSA252) SAOUHSC_02478 (rplM) 50S ribosomal protein L13 [3] (data from MRSA252) SAOUHSC_02492 (rplO) 50S ribosomal protein L15 [3] (data from MRSA252) SAOUHSC_02495 (rplR) 50S ribosomal protein L18 [3] (data from MRSA252) SAOUHSC_01211 (rplS) 50S ribosomal protein L19 [3] (data from MRSA252) SAOUHSC_01784 (rplT) 50S ribosomal protein L20 [3] (data from MRSA252) SAOUHSC_01757 (rplU) 50S ribosomal protein L21 [3] (data from MRSA252) SAOUHSC_02507 (rplV) 50S ribosomal protein L22 [3] (data from MRSA252) SAOUHSC_02510 (rplW) 50S ribosomal protein L23 [3] (data from MRSA252) SAOUHSC_02501 (rplX) 50S ribosomal protein L24 [3] (data from MRSA252) SAOUHSC_01755 (rpmA) 50S ribosomal protein L27 [3] (data from MRSA252) SAOUHSC_01191 (rpmB) 50S ribosomal protein L28 [3] (data from MRSA252) SAOUHSC_02361 (rpmE2) 50S ribosomal protein L31 type B [3] (data from MRSA252) SAOUHSC_01785 (rpmI) 50S ribosomal protein L35 [3] (data from MRSA252) SAOUHSC_01232 (rpsB) 30S ribosomal protein S2 [3] (data from MRSA252) SAOUHSC_02506 (rpsC) 30S ribosomal protein S3 [3] (data from MRSA252) SAOUHSC_01829 (rpsD) 30S ribosomal protein S4 [3] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_00348 (rpsF) 30S ribosomal protein S6 [3] (data from MRSA252) SAOUHSC_02477 (rpsI) 30S ribosomal protein S9 [3] (data from MRSA252) SAOUHSC_02487 (rpsM) 30S ribosomal protein S13 [3] (data from MRSA252) SAOUHSC_02503 (rpsQ) 30S ribosomal protein S17 [3] (data from MRSA252) SAOUHSC_00350 (rpsR) 30S ribosomal protein S18 [3] (data from MRSA252) SAOUHSC_02508 (rpsS) 30S ribosomal protein S19 [3] (data from MRSA252) SAOUHSC_01678 (rpsU) 30S ribosomal protein S21 [3] (data from MRSA252) SAOUHSC_01216 (sucC) succinyl-CoA synthetase subunit beta [3] (data from MRSA252) SAOUHSC_01788 (thrS) threonyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01234 (tsf) elongation factor Ts [3] (data from MRSA252) SAOUHSC_02353 (upp) uracil phosphoribosyltransferase [3] (data from MRSA252) SAOUHSC_00187 formate acetyltransferase [3] (data from MRSA252) SAOUHSC_00204 globin domain-containing protein [3] (data from MRSA252) SAOUHSC_00206 L-lactate dehydrogenase [3] (data from MRSA252) SAOUHSC_00220 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [3] (data from MRSA252) SAOUHSC_00284 5'-nucleotidase [3] (data from MRSA252) SAOUHSC_00336 acetyl-CoA acyltransferase [3] (data from MRSA252) SAOUHSC_00444 hypothetical protein [3] (data from MRSA252) SAOUHSC_00467 pur operon repressor [3] (data from MRSA252) SAOUHSC_00469 regulatory protein SpoVG [3] (data from MRSA252) SAOUHSC_00472 ribose-phosphate pyrophosphokinase [3] (data from MRSA252) SAOUHSC_00483 hypothetical protein [3] (data from MRSA252) SAOUHSC_00485 hypoxanthine phosphoribosyltransferase [3] (data from MRSA252) SAOUHSC_00488 hypothetical protein [3] (data from MRSA252) SAOUHSC_00525 DNA-directed RNA polymerase subunit beta' [3] (data from MRSA252) SAOUHSC_00528 30S ribosomal protein S7 [3] (data from MRSA252) SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_00532 2-amino-3-ketobutyrate coenzyme A ligase [3] (data from MRSA252) SAOUHSC_00535 hypothetical protein [3] (data from MRSA252) SAOUHSC_00536 branched-chain amino acid aminotransferase [3] (data from MRSA252) SAOUHSC_00634 ABC transporter substrate-binding protein [3] (data from MRSA252) SAOUHSC_00655 dihydroxyacetone kinase subunit DhaK [3] (data from MRSA252) SAOUHSC_00679 hypothetical protein [3] (data from MRSA252) SAOUHSC_00690 hypothetical protein [3] (data from MRSA252) SAOUHSC_00795 glyceraldehyde-3-phosphate dehydrogenase [3] (data from MRSA252) SAOUHSC_00865 hypothetical protein [3] (data from MRSA252) SAOUHSC_00920 3-oxoacyl-ACP synthase III [3] (data from MRSA252) SAOUHSC_00937 oligoendopeptidase F [3] (data from MRSA252) SAOUHSC_00947 enoyl-(acyl carrier protein) reductase [3] (data from MRSA252) SAOUHSC_00951 hypothetical protein [3] (data from MRSA252) SAOUHSC_00985 1,4-dihydroxy-2-naphthoyl-CoA synthase [3] (data from MRSA252) SAOUHSC_01029 phosphoenolpyruvate-protein phosphotransferase [3] (data from MRSA252) SAOUHSC_01035 hypothetical protein [3] (data from MRSA252) SAOUHSC_01149 cell division protein [3] (data from MRSA252) SAOUHSC_01150 cell division protein FtsZ [3] (data from MRSA252) SAOUHSC_01154 hypothetical protein [3] (data from MRSA252) SAOUHSC_01193 hypothetical protein [3] (data from MRSA252) SAOUHSC_01198 malonyl CoA-acyl carrier protein transacylase [3] (data from MRSA252) SAOUHSC_01240 prolyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01251 polynucleotide phosphorylase/polyadenylase [3] (data from MRSA252) SAOUHSC_01327 catalase [3] (data from MRSA252) SAOUHSC_01337 transketolase [3] (data from MRSA252) SAOUHSC_01351 DNA topoisomerase IV subunit B [3] (data from MRSA252) SAOUHSC_01431 methionine sulfoxide reductase B [3] (data from MRSA252) SAOUHSC_01490 DNA-binding protein HU [3] (data from MRSA252) SAOUHSC_01626 proline dipeptidase [3] (data from MRSA252) SAOUHSC_01632 glycine dehydrogenase subunit 2 [3] (data from MRSA252) SAOUHSC_01653 superoxide dismutase [3] (data from MRSA252) SAOUHSC_01659 hypothetical protein [3] (data from MRSA252) SAOUHSC_01698 hypothetical protein [3] (data from MRSA252) SAOUHSC_01719 hypothetical protein [3] (data from MRSA252) SAOUHSC_01806 pyruvate kinase [3] (data from MRSA252) SAOUHSC_01807 6-phosphofructokinase [3] (data from MRSA252) SAOUHSC_01814 hypothetical protein [3] (data from MRSA252) SAOUHSC_01819 hypothetical protein [3] (data from MRSA252) SAOUHSC_01838 hypothetical protein [3] (data from MRSA252) SAOUHSC_01845 formate--tetrahydrofolate ligase [3] (data from MRSA252) SAOUHSC_01850 catabolite control protein A [3] (data from MRSA252) SAOUHSC_01852 bifunctional 3-deoxy-7-phosphoheptulonate synthase/chorismate mutase [3] (data from MRSA252) SAOUHSC_01854 hypothetical protein [3] (data from MRSA252) SAOUHSC_01858 hypothetical protein [3] (data from MRSA252) SAOUHSC_01860 hypothetical protein [3] (data from MRSA252) SAOUHSC_01868 dipeptidase PepV [3] (data from MRSA252) SAOUHSC_01901 putative translaldolase [3] (data from MRSA252) SAOUHSC_01910 phosphoenolpyruvate carboxykinase [3] (data from MRSA252) SAOUHSC_02150 hypothetical protein [3] (data from MRSA252) SAOUHSC_02300 STAS domain-containing protein [3] (data from MRSA252) SAOUHSC_02341 F0F1 ATP synthase subunit beta [3] (data from MRSA252) SAOUHSC_02365 UDP-N-acetylglucosamine 1-carboxyvinyltransferase [3] (data from MRSA252) SAOUHSC_02377 pyrimidine-nucleoside phosphorylase [3] (data from MRSA252) SAOUHSC_02399 glucosamine--fructose-6-phosphate aminotransferase [3] (data from MRSA252) SAOUHSC_02403 mannitol-1-phosphate 5-dehydrogenase [3] (data from MRSA252) SAOUHSC_02485 DNA-directed RNA polymerase subunit alpha [3] (data from MRSA252) SAOUHSC_02486 30S ribosomal protein S11 [3] (data from MRSA252) SAOUHSC_02504 50S ribosomal protein L29 [3] (data from MRSA252) SAOUHSC_02577 D-isomer specific 2-hydroxyacid dehydrogenase NAD binding domain-containing protein [3] (data from MRSA252) SAOUHSC_02869 1-pyrroline-5-carboxylate dehydrogenase [3] (data from MRSA252) SAOUHSC_02887 immunodominant antigen A [3] (data from MRSA252) SAOUHSC_02969 arginine deiminase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
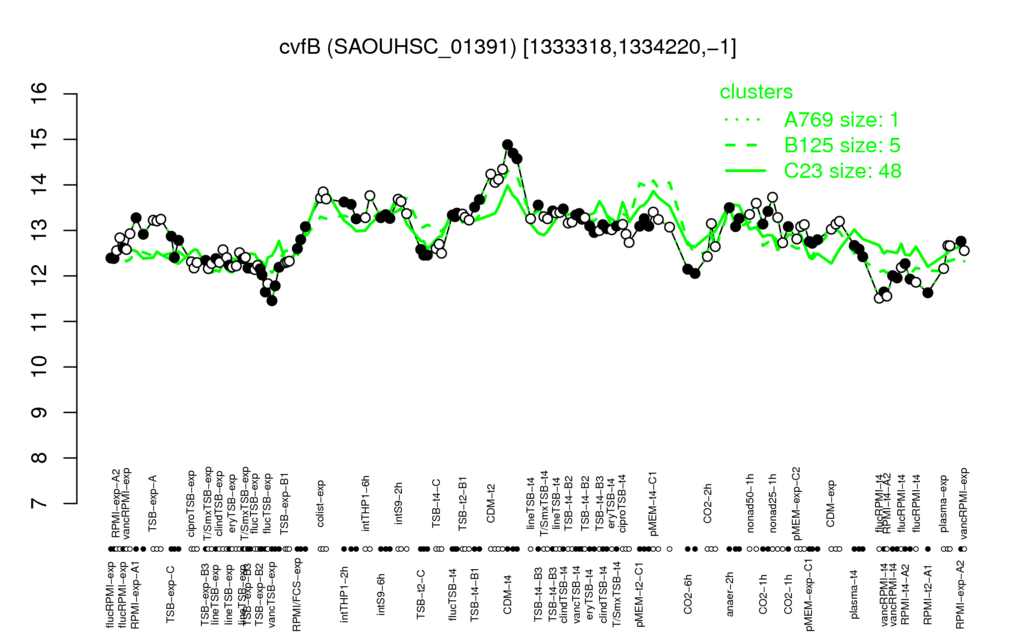 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.000 3.001 3.002 3.003 3.004 3.005 3.006 3.007 3.008 3.009 3.010 3.011 3.012 3.013 3.014 3.015 3.016 3.017 3.018 3.019 3.020 3.021 3.022 3.023 3.024 3.025 3.026 3.027 3.028 3.029 3.030 3.031 3.032 3.033 3.034 3.035 3.036 3.037 3.038 3.039 3.040 3.041 3.042 3.043 3.044 3.045 3.046 3.047 3.048 3.049 3.050 3.051 3.052 3.053 3.054 3.055 3.056 3.057 3.058 3.059 3.060 3.061 3.062 3.063 3.064 3.065 3.066 3.067 3.068 3.069 3.070 3.071 3.072 3.073 3.074 3.075 3.076 3.077 3.078 3.079 3.080 3.081 3.082 3.083 3.084 3.085 3.086 3.087 3.088 3.089 3.090 3.091 3.092 3.093 3.094 3.095 3.096 3.097 3.098 3.099 3.100 3.101 3.102 3.103 3.104 3.105 3.106 3.107 3.108 3.109 3.110 3.111 3.112 3.113 3.114 3.115 3.116 3.117 3.118 3.119 3.120 3.121 3.122 3.123 3.124 3.125 3.126 3.127 3.128 3.129 3.130 3.131 3.132 3.133 3.134 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Bret M Benton, J P Zhang, Skip Bond, Casey Pope, Todd Christian, Lawrence Lee, Kelly M Winterberg, Molly B Schmid, Jerry M Buysse
Large-scale identification of genes required for full virulence of Staphylococcus aureus.
J Bacteriol: 2004, 186(24);8478-89
[PubMed:15576798] [WorldCat.org] [DOI] (P p)Chikara Kaito, Kenji Kurokawa, Yasuhiko Matsumoto, Yutaka Terao, Shigetada Kawabata, Shigeyuki Hamada, Kazuhisa Sekimizu
Silkworm pathogenic bacteria infection model for identification of novel virulence genes.
Mol Microbiol: 2005, 56(4);934-44
[PubMed:15853881] [WorldCat.org] [DOI] (P p)Yasuhiko Matsumoto, Chikara Kaito, Daisuke Morishita, Kenji Kurokawa, Kazuhisa Sekimizu
Regulation of exoprotein gene expression by the Staphylococcus aureus cvfB gene.
Infect Immun: 2007, 75(4);1964-72
[PubMed:17283102] [WorldCat.org] [DOI] (P p)
