Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00187
- pan locus tag?: SAUPAN001082000
- symbol: SAOUHSC_00187
- pan gene symbol?: pflB
- synonym:
- product: formate acetyltransferase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00187
- symbol: SAOUHSC_00187
- product: formate acetyltransferase
- replicon: chromosome
- strand: +
- coordinates: 205908..208157
- length: 2250
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919501 NCBI
- RefSeq: YP_498784 NCBI
- BioCyc: G1I0R-175 BioCyc
- MicrobesOnline: 1288678 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221ATGTTAGAAACAAATAAAAATCATGCAACAGCTTGGCAAGGATTTAAAAATGGAAGATGG
AACAGACACGTAGATGTAAGAGAGTTTATCCAATTAAACTACACTCTTTATGAAGGTAAT
GATTCATTTTTAGCAGGACCAACAGAAGCAACTTCTAAACTTTGGGAACAAGTAATGCAG
TTATCGAAAGAAGAACGTGAACGTGGCGGCATGTGGGATATGGACACGAAAGTAGCTTCA
ACAATCACATCTCATGATGCTGGTTATTTAGACAAAGATTTAGAAACAATTGTAGGTGTA
CAAACTGAAAAGCCATTCAAACGTTCAATGCAACCATTCGGTGGTATTCGTATGGCGAAA
GCAGCTTGTGAAGCTTACGGTTACGAATTAGACGAAGAAACTGAAAAAATCTTTACAGAT
TATCGTAAAACACATAACCAAGGTGTATTCGATGCATATTCTAGAGAAATGTTGAACTGC
CGTAAAGCAGGTGTAATCACTGGTTTACCTGATGCATACGGACGTGGACGTATTATCGGT
GACTATCGTCGTGTAGCTTTATATGGTGTAGATTTCTTAATGGAAGAAAAAATGCACGAC
TTCAACACGATGTCTACAGAAATGTCAGAAGATGTAATTCGTTTACGTGAAGAATTATCA
GAACAATATCGTGCATTAAAAGAATTAAAAGAACTTGGACAAAAATATGGTTTCGATTTA
AGCCGTCCAGCAGAAAACTTCAAAGAAGCAGTTCAATGGTTATACTTAGCATACCTTGCT
GCAATTAAAGAACAAAACGGTGCAGCAATGAGTTTAGGTCGTACATCAACATTCTTAGAT
ATCTATGCTGAACGTGACCTTAAAGCAGGCGTTATTACTGAAAGCGAAGTTCAAGAAATT
ATTGACCACTTCATCATGAAATTACGTATTGTTAAATTTGCTCGTACACCTGATTACAAT
GAATTATTCTCTGGAGACCCAACTTGGGTAACTGAATCTATCGGTGGTGTAGGTATTGAC
GGACGTCCACTTGTTACGAAAAACTCATTCCGTTTCTTACACTCATTAGATAACTTAGGT
CCAGCTCCAGAACCAAACTTAACAGTATTATGGTCAGTACGTTTACCTGACAACTTCAAA
ACATACTGTGCAAAAATGAGTATTAAAACAAGTTCTATCCAATATGAAAATGATGACATT
ATGCGTGAAAGCTATGGCGATGACTATGGTATCGCATGTTGTGTATCAGCGATGACAATT
GGTAAACAAATGCAATTCTTCGGTGCACGTGCGAACTTAGCTAAAACATTACTTTACGCT
ATCAATGGTGGTAAAGATGAAAAATCTGGTGCACAAGTTGGTCCAAACTTCGAAGGTATT
AACAGCGAAGTATTAGAATATGACGAAGTATTCAAGAAATTTGATCAAATGATGGATTGG
CTAGCAGGTGTTTACATTAACTCATTAAATGTTATTCACTACATGCACGATAAATACAGC
TATGAACGTATTGAAATGGCATTACATGATACAGAAATTGTACGTACAATGGCAACAGGT
ATCGCTGGTTTATCAGTAGCAGCTGACTCATTATCTGCAATTAAATATGCACAAGTTAAA
CCAATTCGTAACGAAGAAGGTCTTGTAGTAGACTTTGAAATCGAAGGCGACTTCCCTAAA
TACGGTAACAATGACGACCGTGTAGATGATATTGCAGTTGATTTAGTAGAACGCTTCATG
ACTAAATTACGTAGTCATAAAACATATCGTGATTCAGAACATACAATGAGTGTATTAACA
ATTACTTCAAACGTTGTATACGGTAAGAAAACTGGTAACACACCAGACGGACGTAAAGCT
GGCGAACCATTTGCTCCAGGTGCAAACCCAATGCATGGCCGTGACCAAAAAGGTGCATTA
TCTTCATTAAGTTCTGTAGCTAAGATCCCTTACGATTGCTGTAAAGATGGTATTTCAAAT
ACATTCAGTATCGTACCAAAATCATTAGGTAAAGAACCAGAAGATCAAAACCGTAACTTA
ACTAGTATGTTAGATGGTTACGCAATGCAATGTGGTCACCACTTAAATATTAACGTATTT
AACCGTGAAACATTAATAGATGCAATGGAACATCCAGAAGAATATCCACAGTTAACAATC
CGTGTATCTGGTTACGCTGTTAACTTCATTAAATTAACACGTGAACAACAATTAGATGTA
ATTTCTCGTACATTCCATGAAAGTATGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2250
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00187
- symbol: SAOUHSC_00187
- description: formate acetyltransferase
- length: 749
- theoretical pI: 5.14561
- theoretical MW: 84861.4
- GRAVY: -0.470227
⊟Function[edit | edit source]
- reaction: EC 2.3.1.54? ExPASyFormate C-acetyltransferase Acetyl-CoA + formate = CoA + pyruvate
- TIGRFAM: Energy metabolism Fermentation formate acetyltransferase (TIGR01255; EC 2.3.1.54; HMM-score: 1268.1)and 3 moreglycyl radical enzyme, PFL2/glycerol dehydratase family (TIGR01774; HMM-score: 239.7)Energy metabolism Amino acids and amines choline trimethylamine-lyase (TIGR04394; EC 4.3.99.4; HMM-score: 139.6)autonomous glycyl radical cofactor GrcA (TIGR04365; HMM-score: 89.5)
- TheSEED :
- Pyruvate formate-lyase (EC 2.3.1.54)
and 2 more - PFAM: PFL-like (CL0339) PFL-like; Pyruvate formate lyase-like (PF02901; HMM-score: 745)and 2 moreGly_radical; Glycine radical (PF01228; HMM-score: 134.4)YjjI-like; Glycyl radical enzyme YjjI-like (PF11230; HMM-score: 21.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 10
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9352
- Cytoplasmic Membrane Score: 0.0015
- Cell wall & surface Score: 0.0002
- Extracellular Score: 0.0631
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003741
- TAT(Tat/SPI): 0.000313
- LIPO(Sec/SPII): 0.001076
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLETNKNHATAWQGFKNGRWNRHVDVREFIQLNYTLYEGNDSFLAGPTEATSKLWEQVMQLSKEERERGGMWDMDTKVASTITSHDAGYLDKDLETIVGVQTEKPFKRSMQPFGGIRMAKAACEAYGYELDEETEKIFTDYRKTHNQGVFDAYSREMLNCRKAGVITGLPDAYGRGRIIGDYRRVALYGVDFLMEEKMHDFNTMSTEMSEDVIRLREELSEQYRALKELKELGQKYGFDLSRPAENFKEAVQWLYLAYLAAIKEQNGAAMSLGRTSTFLDIYAERDLKAGVITESEVQEIIDHFIMKLRIVKFARTPDYNELFSGDPTWVTESIGGVGIDGRPLVTKNSFRFLHSLDNLGPAPEPNLTVLWSVRLPDNFKTYCAKMSIKTSSIQYENDDIMRESYGDDYGIACCVSAMTIGKQMQFFGARANLAKTLLYAINGGKDEKSGAQVGPNFEGINSEVLEYDEVFKKFDQMMDWLAGVYINSLNVIHYMHDKYSYERIEMALHDTEIVRTMATGIAGLSVAADSLSAIKYAQVKPIRNEEGLVVDFEIEGDFPKYGNNDDRVDDIAVDLVERFMTKLRSHKTYRDSEHTMSVLTITSNVVYGKKTGNTPDGRKAGEPFAPGANPMHGRDQKGALSSLSSVAKIPYDCCKDGISNTFSIVPKSLGKEPEDQNRNLTSMLDGYAMQCGHHLNINVFNRETLIDAMEHPEEYPQLTIRVSGYAVNFIKLTREQQLDVISRTFHESM
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_00679 hypothetical protein [3] (data from MRSA252) SAOUHSC_01451 threonine dehydratase [3] (data from MRSA252) SAOUHSC_01819 hypothetical protein [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulators: CcpA* regulon, Rex* (repression) regulon
CcpA* (TF) important in Carbon catabolism; RegPrecise Rex* (TF) important in Energy metabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
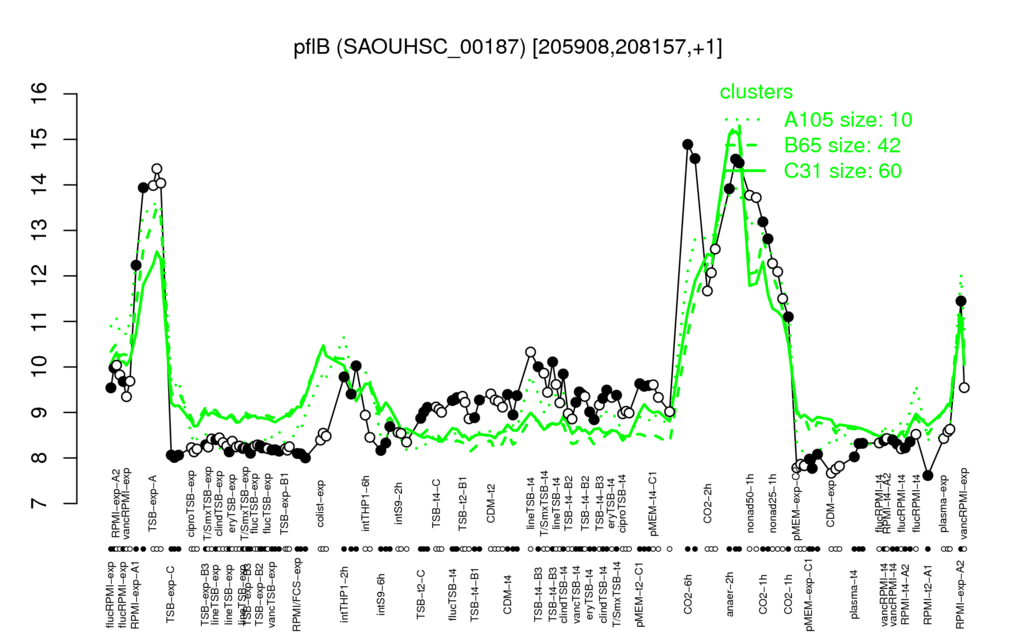 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 3.3 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Martina Leibig, Manuel Liebeke, Diana Mader, Michael Lalk, Andreas Peschel, Friedrich Götz
Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus.
J Bacteriol: 2011, 193(4);952-62
[PubMed:21169491] [WorldCat.org] [DOI] (I p)
