Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01829
- pan locus tag?: SAUPAN004372000
- symbol: rpsD
- pan gene symbol?: rpsD
- synonym:
- product: 30S ribosomal protein S4
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3921779 NCBI
- RefSeq: YP_500334 NCBI
- BioCyc: G1I0R-1701 BioCyc
- MicrobesOnline: 1290248 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601ATGGCTCGATTCAGAGGTTCAAACTGGAAAAAATCTCGTCGTTTAGGTATCTCTTTAAGC
GGTACTGGTAAAGAATTAGAAAAACGTCCTTACGCACCAGGACAACATGGTCCAAACCAA
CGTAAAAAATTATCAGAATATGGTTTACAATTACGTGAAAAACAAAAATTACGTTACTTA
TATGGAATGACTGAAAGACAATTCCGTAACACATTTGACATCGCTGGTAAAAAATTCGGT
GTACACGGTGAAAACTTCATGATCTTATTAGCAAGTCGTTTAGACGCTGTTGTTTATTCA
TTAGGTTTAGCTCGTACTCGTCGTCAAGCACGTCAATTAGTTAACCACGGTCATATCTTA
GTAGATGGTAAACGTGTTGATATTCCATCTTATTCTGTTAAACCTGGTCAAACAATTTCA
GTTCGTGAAAAATCTCAAAAATTAAACATCATCGTTGAATCAGTTGAAATCAACAATTTC
GTACCTGAGTACTTAAACTTTGATGCTGACAGCTTAACTGGTACTTTCGTACGTTTACCA
GAACGTAGCGAATTACCTGCTGAAATTAACGAACAATTAATCGTTGAGTACTACTCAAGA
TAA60
120
180
240
300
360
420
480
540
600
603
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01829
- symbol: RpsD
- description: 30S ribosomal protein S4
- length: 200
- theoretical pI: 10.559
- theoretical MW: 23013.2
- GRAVY: -0.607
⊟Function[edit | edit source]
- TIGRFAM: Protein synthesis Ribosomal proteins: synthesis and modification ribosomal protein uS4 (TIGR01017; HMM-score: 271.5)and 4 moreProtein synthesis Ribosomal proteins: synthesis and modification ribosomal protein uS4 (TIGR01018; HMM-score: 52.7)Unknown function General TlyA family rRNA methyltransferase/putative hemolysin (TIGR00478; HMM-score: 28)Protein synthesis tRNA and rRNA base modification pseudouridine synthase, RluA family (TIGR00005; EC 5.4.99.-; HMM-score: 14.8)Energy metabolism Photosynthesis photosystem II S4 domain protein (TIGR03069; HMM-score: 14.8)
- TheSEED :
- SSU ribosomal protein S4p (S9e)
- SSU ribosomal protein S4p (S9e), zinc-independent
- PFAM: S4 (CL0492) Ribosomal_S4; Ribosomal protein S4/S9 N-terminal domain (PF00163; HMM-score: 90.1)S4; S4 domain (PF01479; HMM-score: 73.5)and 1 moreSYY_C-terminal; Tyrosine--tRNA ligase SYY-like C-terminal domain (PF22421; HMM-score: 20.9)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0.2528
- Cytoplasmic Membrane Score: 0.0013
- Cell wall & surface Score: 0
- Extracellular Score: 0.7459
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.016333
- TAT(Tat/SPI): 0.011778
- LIPO(Sec/SPII): 0.010986
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MARFRGSNWKKSRRLGISLSGTGKELEKRPYAPGQHGPNQRKKLSEYGLQLREKQKLRYLYGMTERQFRNTFDIAGKKFGVHGENFMILLASRLDAVVYSLGLARTRRQARQLVNHGHILVDGKRVDIPSYSVKPGQTISVREKSQKLNIIVESVEINNFVPEYLNFDADSLTGTFVRLPERSELPAEINEQLIVEYYSR
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_02380 (deoD) purine nucleoside phosphorylase [4] (data from MRSA252) SAOUHSC_01683 (dnaK) molecular chaperone DnaK [4] (data from MRSA252) SAOUHSC_00799 (eno) phosphopyruvate hydratase [4] (data from MRSA252) SAOUHSC_01786 (infC) translation initiation factor IF-3 [4] (data from MRSA252) SAOUHSC_00900 (pgi) glucose-6-phosphate isomerase [4] (data from MRSA252) SAOUHSC_00519 (rplA) 50S ribosomal protein L1 [4] (data from MRSA252) SAOUHSC_02509 (rplB) 50S ribosomal protein L2 [4] (data from MRSA252) SAOUHSC_02512 (rplC) 50S ribosomal protein L3 [4] (data from MRSA252) SAOUHSC_02511 (rplD) 50S ribosomal protein L4 [4] (data from MRSA252) SAOUHSC_00017 (rplI) 50S ribosomal protein L9 [4] (data from MRSA252) SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [4] (data from MRSA252) SAOUHSC_00518 (rplK) 50S ribosomal protein L11 [4] (data from MRSA252) SAOUHSC_00521 (rplL) 50S ribosomal protein L7/L12 [4] (data from MRSA252) SAOUHSC_02492 (rplO) 50S ribosomal protein L15 [4] (data from MRSA252) SAOUHSC_02505 (rplP) 50S ribosomal protein L16 [4] (data from MRSA252) SAOUHSC_02484 (rplQ) 50S ribosomal protein L17 [4] (data from MRSA252) SAOUHSC_01211 (rplS) 50S ribosomal protein L19 [4] (data from MRSA252) SAOUHSC_01784 (rplT) 50S ribosomal protein L20 [4] (data from MRSA252) SAOUHSC_01757 (rplU) 50S ribosomal protein L21 [4] (data from MRSA252) SAOUHSC_02507 (rplV) 50S ribosomal protein L22 [4] (data from MRSA252) SAOUHSC_02510 (rplW) 50S ribosomal protein L23 [4] (data from MRSA252) SAOUHSC_02501 (rplX) 50S ribosomal protein L24 [4] (data from MRSA252) SAOUHSC_01755 (rpmA) 50S ribosomal protein L27 [4] (data from MRSA252) SAOUHSC_02361 (rpmE2) 50S ribosomal protein L31 type B [4] (data from MRSA252) SAOUHSC_01785 (rpmI) 50S ribosomal protein L35 [4] (data from MRSA252) SAOUHSC_00524 (rpoB) DNA-directed RNA polymerase subunit beta [4] (data from MRSA252) SAOUHSC_02506 (rpsC) 30S ribosomal protein S3 [4] (data from MRSA252) SAOUHSC_00348 (rpsF) 30S ribosomal protein S6 [4] (data from MRSA252) SAOUHSC_02498 (rpsH) 30S ribosomal protein S8 [4] (data from MRSA252) SAOUHSC_02477 (rpsI) 30S ribosomal protein S9 [4] (data from MRSA252) SAOUHSC_00527 (rpsL) 30S ribosomal protein S12 [4] (data from MRSA252) SAOUHSC_02487 (rpsM) 30S ribosomal protein S13 [4] (data from MRSA252) SAOUHSC_01250 (rpsO) 30S ribosomal protein S15 [4] (data from MRSA252) SAOUHSC_02503 (rpsQ) 30S ribosomal protein S17 [4] (data from MRSA252) SAOUHSC_02508 (rpsS) 30S ribosomal protein S19 [4] (data from MRSA252) SAOUHSC_00187 formate acetyltransferase [4] (data from MRSA252) SAOUHSC_00206 L-lactate dehydrogenase [4] (data from MRSA252) SAOUHSC_00284 5'-nucleotidase [4] (data from MRSA252) SAOUHSC_00444 hypothetical protein [4] (data from MRSA252) SAOUHSC_00474 50S ribosomal protein L25/general stress protein Ctc [4] (data from MRSA252) SAOUHSC_00528 30S ribosomal protein S7 [4] (data from MRSA252) SAOUHSC_00679 hypothetical protein [4] (data from MRSA252) SAOUHSC_00690 hypothetical protein [4] (data from MRSA252) SAOUHSC_00742 ribonucleotide-diphosphate reductase subunit alpha [4] (data from MRSA252) SAOUHSC_00767 hypothetical protein [4] (data from MRSA252) SAOUHSC_00795 glyceraldehyde-3-phosphate dehydrogenase [4] (data from MRSA252) SAOUHSC_00798 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [4] (data from MRSA252) SAOUHSC_00835 hypothetical protein [4] (data from MRSA252) SAOUHSC_00878 hypothetical protein [4] (data from MRSA252) SAOUHSC_00895 glutamate dehydrogenase [4] (data from MRSA252) SAOUHSC_01035 hypothetical protein [4] (data from MRSA252) SAOUHSC_01040 pyruvate dehydrogenase complex, E1 component subunit alpha [4] (data from MRSA252) SAOUHSC_01199 3-oxoacyl-(acyl-carrier-protein) reductase [4] (data from MRSA252) SAOUHSC_01228 transcriptional repressor CodY [4] (data from MRSA252) SAOUHSC_01287 glutamine synthetase [4] (data from MRSA252) SAOUHSC_01337 transketolase [4] (data from MRSA252) SAOUHSC_01391 hypothetical protein [4] (data from MRSA252) SAOUHSC_01490 DNA-binding protein HU [4] (data from MRSA252) SAOUHSC_01626 proline dipeptidase [4] (data from MRSA252) SAOUHSC_01698 hypothetical protein [4] (data from MRSA252) SAOUHSC_01801 isocitrate dehydrogenase [4] (data from MRSA252) SAOUHSC_01814 hypothetical protein [4] (data from MRSA252) SAOUHSC_01838 hypothetical protein [4] (data from MRSA252) SAOUHSC_01854 hypothetical protein [4] (data from MRSA252) SAOUHSC_02140 manganese-dependent inorganic pyrophosphatase [4] (data from MRSA252) SAOUHSC_02316 DEAD-box ATP dependent DNA helicase [4] (data from MRSA252) SAOUHSC_02366 fructose-bisphosphate aldolase [4] (data from MRSA252) SAOUHSC_02369 DNA-directed RNA polymerase subunit delta [4] (data from MRSA252) SAOUHSC_02377 pyrimidine-nucleoside phosphorylase [4] (data from MRSA252) SAOUHSC_02399 glucosamine--fructose-6-phosphate aminotransferase [4] (data from MRSA252) SAOUHSC_02441 alkaline shock protein 23 [4] (data from MRSA252) SAOUHSC_02486 30S ribosomal protein S11 [4] (data from MRSA252) SAOUHSC_02504 50S ribosomal protein L29 [4] (data from MRSA252) SAOUHSC_02527 peptidoglycan pentaglycine interpeptide biosynthesis protein FmhB [4] (data from MRSA252) SAOUHSC_02927 malate:quinone oxidoreductase [4] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [5] : SAOUHSC_01828 > S724 > rpsD
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [5]
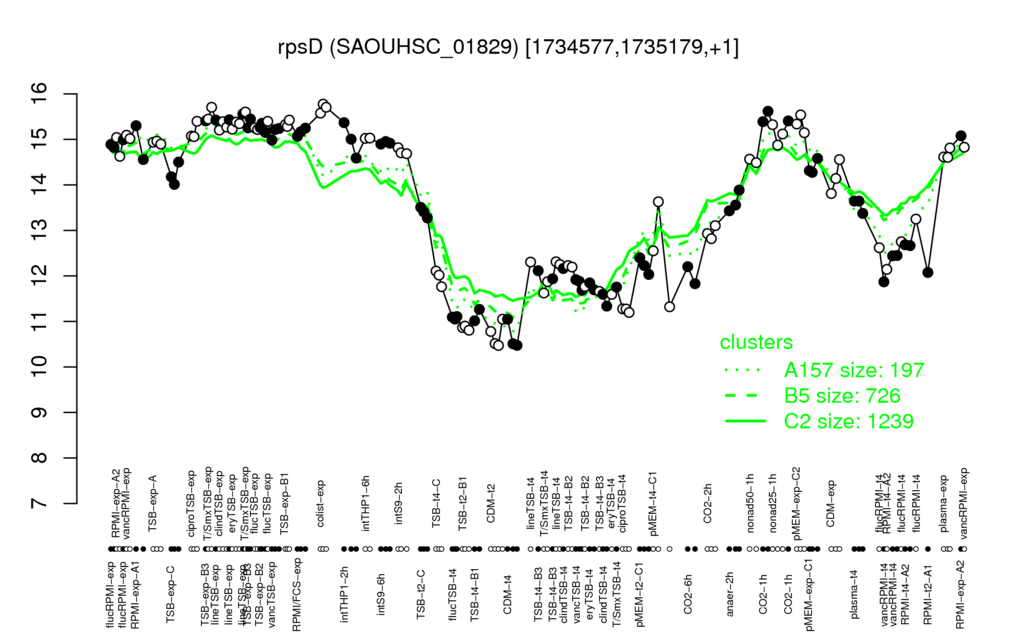 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 4.20 4.21 4.22 4.23 4.24 4.25 4.26 4.27 4.28 4.29 4.30 4.31 4.32 4.33 4.34 4.35 4.36 4.37 4.38 4.39 4.40 4.41 4.42 4.43 4.44 4.45 4.46 4.47 4.48 4.49 4.50 4.51 4.52 4.53 4.54 4.55 4.56 4.57 4.58 4.59 4.60 4.61 4.62 4.63 4.64 4.65 4.66 4.67 4.68 4.69 4.70 4.71 4.72 4.73 4.74 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 5.0 5.1 5.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
