Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL1449 [new locus tag: SACOL_RS07390 ]
- pan locus tag?: SAUPAN003831000
- symbol: sucA
- pan gene symbol?: sucA
- synonym:
- product: 2-oxoglutarate dehydrogenase E1 component
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL1449 [new locus tag: SACOL_RS07390 ]
- symbol: sucA
- product: 2-oxoglutarate dehydrogenase E1 component
- replicon: chromosome
- strand: -
- coordinates: 1460973..1463771
- length: 2799
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3238284 NCBI
- RefSeq: YP_186301 NCBI
- BioCyc: see SACOL_RS07390
- MicrobesOnline: 912907 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761ATGACTAACGAAAGAAAAGAAGTTTCAGAGGCTCCTGTAAACTTCGGTGCGAATTTAGGT
CTAATGTTAGATCTATATGATGACTTTTTACAAGATCCATCATCTGTACCAGAAGATTTA
CAAGTCTTATTCAGCACAATTAAGAATGATGACTCAATTGTACCAGCTTTAAAAAGTACA
AGTAGTCAAAATAGCGACGGCACAATTAAGCGTGTCATGCGTTTAATTGATAATATTCGC
CAATACGGGCATCTTAAAGCCGATATTTATCCTGTAAATCCTCCAAAAAGGAAACATGTA
CCTAAATTAGAGATTGAAGACTTTGATTTAGATCAACAGACTTTGGAAGGTATATCAGCA
GGAATTGTTTCAGATCACTTTGCCGACATTTATGATAATGCTTATGAAGCAATTTTAAGA
ATGGAAAAACGTTACAAAGGACCAATTGCATTTGAGTATACACATATTAATAACAATACC
GAACGTGGTTGGTTAAAAAGAAGAATTGAAACGCCATATAAAGTAACGTTAAATAATAAC
GAAAAAAGGGCACTATTCAAACAATTAGCGTATGTTGAAGGGTTTGAAAAATATCTTCAT
AAAAACTTCGTTGGTGCAAAGCGTTTTTCAATTGAAGGGGTAGACGCACTTGTACCGATG
TTACAACGTACTATTACGATTGCTGCGAAAGAAGGTATTAAAAATATACAAATAGGCATG
GCTCACCGTGGACGTTTAAACGTTTTAACGCATGTCTTAGAAAAACCGTACGAAATGATG
ATTTCAGAATTTATGCATACAGATCCAATGAAATTCTTACCTGAAGATGGTAGCTTGCAG
TTAACTGCTGGATGGACTGGTGATGTGAAATATCACCTTGGTGGCATTAAAACTACTGAT
TCATACGGTACAATGCAGCGTATTGCACTGGCTAACAATCCAAGTCACTTGGAAATTGTT
GCACCTGTTGTTGAGGGGCGTACGAGAGCAGCACAAGATGATACACAACGAGCTGGGGCT
CCGACGACTGATCATCATAAAGCAATGCCAATTATTATACATGGCGATGCTGCTTATCCT
GGTCAAGGAATTAACTTCGAAACAATGAACTTAGGAAACTTGAAAGGCTATTCTACGGGT
GGTTCATTGCATATTATTACTAACAATAGAATTGGATTTACTACAGAACCAATTGATGCA
CGTTCAACAACTTATTCTACAGATGTGGCCAAAGGTTATGATGTGCCAATATTCCATGTC
AATGCAGATGACGTTGAAGCTACTATTGAAGCAATTGATATTGCAATGGAATTTAGAAAA
GAGTTTCATAAAGACGTCGTTATTGATTTAGTAGGTTATCGTCGTTTCGGACATAACGAA
ATGGATGAACCATCAATTACTAATCCAGTTCCTTATCAGAATATTCGCAAACATGACTCT
GTTGAATATGTGTTTGGTAAAAAGCTTGTTAATGAAGGTGTCATTTCAGAAGATGAAATG
CATTCATTTATAGAACAAGTCCAAAAGGAACTAAGACAAGCTCATGATAAAATTAATAAA
GCTGATAAAATGGATAATCCAGATATGGAAAAGCCTGCAGATCTTGCATTACCGTTACAA
GCAGACGAACAATCATTTACTTTTGATCACTTGAAAGAAATAAATGATGCATTGTTAACA
TATCCGGATGGCTTTAACATTTTGAAAAAGTTAAACAAAGTTCTTGAGAAGCGTCATGAG
CCGTTTAATAAAGAAGATGGTTTAGTTGATTGGGCACAAGCAGAACAACTTGCATTTGCG
ACAATTTTACAAGATGGTACACCGATTCGCTTAACTGGTCAAGATAGTGAACGTGGTACA
TTCAGTCATAGGCATGCCGTGTTACATGATGAGCAAACAGGTGAAACATATACACCTTTA
CATCATGTTCCTGATCAAAAAGCGACATTTGATATACACAATTCTCCGCTTTCAGAAGCA
GCAGTAGTTGGTTTTGAATACGGCTATAATGTGGAAAACAAAAAAAGCTTCAATATTTGG
GAAGCACAATATGGTGATTTTGCAAATATGTCACAAATGATTTTTGACAACTTCTTATTC
AGTTCTCGCTCAAAATGGGGAGAACGTTCAGGATTAACATTATTCTTACCTCATGCATAT
GAGGGTCAAGGGCCTGAACATTCATCAGCAAGATTAGAGCGATTTTTACAATTAGCTGCT
GAAAATAATTGCACAGTTGTCAACTTATCTAGTTCAAGTAATTATTTCCACTTATTGCGT
GCACAAGCGGCTAGTTTAGATTCTGAACAAATGCGACCATTGGTTGTTATGTCACCAAAA
AGCTTACTGAGAAATAAAACAGTTGCAAAACCAATTGATGAATTTACTTCTGGTGGATTT
GAGCCAATTTTGACAGAATCATATCAAGCGGATAAGGTTACAAAAGTTATTTTGGCAACT
GGTAAAATGTTCATTGATTTAAAAGAAGCATTAGCTAAAAATCCAGACGAATCAGTATTA
CTCGTTGCGATTGAAAGATTGTATCCATTCCCAGAGGAAGAGATTGAAGCATTACTAGCA
CAATTGCCAAACCTTGAAGAAGTGTCATGGGTACAAGAAGAACCTAAAAATCAAGGTGCA
TGGTTATATGTCTATCCATATGTTAAAGTGCTAGTTGCAGATAAATATGATTTAAGTTAT
CATGGCAGAATTCAAAGGGCTGCTCCAGCTGAAGGCGATGGAGAAATTCATAAACTTGTT
CAAAATAAAATTATAGAAAATGCATTAAAAAATAACTAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2799
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL1449 [new locus tag: SACOL_RS07390 ]
- symbol: SucA
- description: 2-oxoglutarate dehydrogenase E1 component
- length: 932
- theoretical pI: 5.40096
- theoretical MW: 105342
- GRAVY: -0.469957
⊟Function[edit | edit source]
- reaction: EC 1.2.4.2? ExPASyOxoglutarate dehydrogenase (succinyl-transferring) 2-oxoglutarate + [dihydrolipoyllysine-residue succinyltransferase] lipoyllysine = [dihydrolipoyllysine-residue succinyltransferase] S-succinyldihydrolipoyllysine + CO2
- TIGRFAM: Energy metabolism TCA cycle oxoglutarate dehydrogenase (succinyl-transferring), E1 component (TIGR00239; EC 1.2.4.2; HMM-score: 1033.1)and 2 moreEnergy metabolism Pyruvate dehydrogenase pyruvate dehydrogenase (acetyl-transferring) E1 component, alpha subunit (TIGR03182; EC 1.2.4.1; HMM-score: 52.6)Energy metabolism Pyruvate dehydrogenase pyruvate dehydrogenase (acetyl-transferring) E1 component, alpha subunit (TIGR03181; EC 1.2.4.1; HMM-score: 31.3)
- TheSEED :
- 2-oxoglutarate dehydrogenase E1 component (EC 1.2.4.2)
Carbohydrates Central carbohydrate metabolism Dehydrogenase complexes 2-oxoglutarate dehydrogenase E1 component (EC 1.2.4.2)and 1 more - PFAM: THDP-binding (CL0254) E1_dh; Dehydrogenase E1 component (PF00676; HMM-score: 206.1)and 5 moreTransket_pyr; Transketolase, pyrimidine binding domain (PF02779; HMM-score: 139.8)TKC_like (CL0591) OxoGdeHyase_C; 2-oxoglutarate dehydrogenase C-terminal (PF16870; HMM-score: 117.2)no clan defined 2-oxogl_dehyd_N; 2-oxoglutarate dehydrogenase N-terminus (PF16078; HMM-score: 21)TPR (CL0020) Importin_rep_6; Importin repeat 6 (PF18829; HMM-score: 12.4)TKC_like (CL0591) PFOR_II; Pyruvate:ferredoxin oxidoreductase core domain II (PF17147; HMM-score: 12.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: thiamine diphosphate
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.95
- Cytoplasmic Membrane Score: 0.05
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.7401
- Cytoplasmic Membrane Score: 0.0506
- Cell wall & surface Score: 0.0031
- Extracellular Score: 0.2062
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.002478
- TAT(Tat/SPI): 0.000638
- LIPO(Sec/SPII): 0.000551
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTNERKEVSEAPVNFGANLGLMLDLYDDFLQDPSSVPEDLQVLFSTIKNDDSIVPALKSTSSQNSDGTIKRVMRLIDNIRQYGHLKADIYPVNPPKRKHVPKLEIEDFDLDQQTLEGISAGIVSDHFADIYDNAYEAILRMEKRYKGPIAFEYTHINNNTERGWLKRRIETPYKVTLNNNEKRALFKQLAYVEGFEKYLHKNFVGAKRFSIEGVDALVPMLQRTITIAAKEGIKNIQIGMAHRGRLNVLTHVLEKPYEMMISEFMHTDPMKFLPEDGSLQLTAGWTGDVKYHLGGIKTTDSYGTMQRIALANNPSHLEIVAPVVEGRTRAAQDDTQRAGAPTTDHHKAMPIIIHGDAAYPGQGINFETMNLGNLKGYSTGGSLHIITNNRIGFTTEPIDARSTTYSTDVAKGYDVPIFHVNADDVEATIEAIDIAMEFRKEFHKDVVIDLVGYRRFGHNEMDEPSITNPVPYQNIRKHDSVEYVFGKKLVNEGVISEDEMHSFIEQVQKELRQAHDKINKADKMDNPDMEKPADLALPLQADEQSFTFDHLKEINDALLTYPDGFNILKKLNKVLEKRHEPFNKEDGLVDWAQAEQLAFATILQDGTPIRLTGQDSERGTFSHRHAVLHDEQTGETYTPLHHVPDQKATFDIHNSPLSEAAVVGFEYGYNVENKKSFNIWEAQYGDFANMSQMIFDNFLFSSRSKWGERSGLTLFLPHAYEGQGPEHSSARLERFLQLAAENNCTVVNLSSSSNYFHLLRAQAASLDSEQMRPLVVMSPKSLLRNKTVAKPIDEFTSGGFEPILTESYQADKVTKVILATGKMFIDLKEALAKNPDESVLLVAIERLYPFPEEEIEALLAQLPNLEEVSWVQEEPKNQGAWLYVYPYVKVLVADKYDLSYHGRIQRAAPAEGDGEIHKLVQNKIIENALKNN
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas
- protein localization: Cytoplasmic [1] [2] [3] [4]
- quantitative data / protein copy number per cell: 581 [5]
- interaction partners:
SACOL0452 (ahpC) alkyl hydroperoxide reductase subunit C [6] (data from MRSA252) SACOL0557 (cysK) cysteine synthase [6] (data from MRSA252) SACOL2130 (deoD) purine nucleoside phosphorylase [6] (data from MRSA252) SACOL1637 (dnaK) molecular chaperone DnaK [6] (data from MRSA252) SACOL0842 (eno) phosphopyruvate hydratase [6] (data from MRSA252) SACOL0634 (eutD) phosphotransacetylase [6] (data from MRSA252) SACOL1782 (fhs) formate--tetrahydrofolate ligase [6] (data from MRSA252) SACOL1199 (ftsZ) cell division protein FtsZ [6] (data from MRSA252) SACOL0593 (fusA) elongation factor G [6] (data from MRSA252) SACOL0838 (gapA1) glyceraldehyde 3-phosphate dehydrogenase [6] (data from MRSA252) SACOL1734 (gapA2) glyceraldehyde 3-phosphate dehydrogenase 2 [6] (data from MRSA252) SACOL1961 (gatA) aspartyl/glutamyl-tRNA amidotransferase subunit A [6] (data from MRSA252) SACOL1960 (gatB) aspartyl/glutamyl-tRNA amidotransferase subunit B [6] (data from MRSA252) SACOL1622 (glyS) glycyl-tRNA synthetase [6] (data from MRSA252) SACOL1554 (gnd) 6-phosphogluconate dehydrogenase [6] (data from MRSA252) SACOL2016 (groEL) chaperonin GroEL [6] (data from MRSA252) SACOL0460 (guaB) inosine-5'-monophosphate dehydrogenase [6] (data from MRSA252) SACOL1513 (hup) DNA-binding protein HU [6] (data from MRSA252) SACOL1741 (icd) isocitrate dehydrogenase [6] (data from MRSA252) SACOL1206 (ileS) isoleucyl-tRNA synthetase [6] (data from MRSA252) SACOL1477 (ilvA1) threonine dehydratase [6] (data from MRSA252) SACOL2623 (mqo2) malate:quinone oxidoreductase [6] (data from MRSA252) SACOL1102 (pdhA) pyruvate dehydrogenase complex E1 component subunit alpha [6] (data from MRSA252) SACOL1103 (pdhB) pyruvate dehydrogenase complex E1 component subunit beta [6] (data from MRSA252) SACOL1104 (pdhC) branched-chain alpha-keto acid dehydrogenase E2 [6] (data from MRSA252) SACOL1105 (pdhD) dihydrolipoamide dehydrogenase [6] (data from MRSA252) SACOL0204 (pflB) formate acetyltransferase [6] (data from MRSA252) SACOL0966 (pgi) glucose-6-phosphate isomerase [6] (data from MRSA252) SACOL0839 (pgk) phosphoglycerate kinase [6] (data from MRSA252) SACOL1091 (ptsH) phosphocarrier protein HPr [6] (data from MRSA252) SACOL1745 (pyk) pyruvate kinase [6] (data from MRSA252) SACOL0584 (rplA) 50S ribosomal protein L1 [6] (data from MRSA252) SACOL2236 (rplB) 50S ribosomal protein L2 [6] (data from MRSA252) SACOL2239 (rplC) 50S ribosomal protein L3 [6] (data from MRSA252) SACOL2238 (rplD) 50S ribosomal protein L4 [6] (data from MRSA252) SACOL2227 (rplE) 50S ribosomal protein L5 [6] (data from MRSA252) SACOL2224 (rplF) 50S ribosomal protein L6 [6] (data from MRSA252) SACOL0585 (rplJ) 50S ribosomal protein L10 [6] (data from MRSA252) SACOL0583 (rplK) 50S ribosomal protein L11 [6] (data from MRSA252) SACOL2220 (rplO) 50S ribosomal protein L15 [6] (data from MRSA252) SACOL2212 (rplQ) 50S ribosomal protein L17 [6] (data from MRSA252) SACOL1257 (rplS) 50S ribosomal protein L19 [6] (data from MRSA252) SACOL1725 (rplT) 50S ribosomal protein L20 [6] (data from MRSA252) SACOL1702 (rplU) 50S ribosomal protein L21 [6] (data from MRSA252) SACOL2234 (rplV) 50S ribosomal protein L22 [6] (data from MRSA252) SACOL2237 (rplW) 50S ribosomal protein L23 [6] (data from MRSA252) SACOL0545 (rplY) 50S ribosomal protein L25/general stress protein Ctc [6] (data from MRSA252) SACOL0588 (rpoB) DNA-directed RNA polymerase subunit beta [6] (data from MRSA252) SACOL1274 (rpsB) 30S ribosomal protein S2 [6] (data from MRSA252) SACOL2233 (rpsC) 30S ribosomal protein S3 [6] (data from MRSA252) SACOL1769 (rpsD) 30S ribosomal protein S4 [6] (data from MRSA252) SACOL2222 (rpsE) 30S ribosomal protein S5 [6] (data from MRSA252) SACOL0437 (rpsF) 30S ribosomal protein S6 [6] (data from MRSA252) SACOL0592 (rpsG) 30S ribosomal protein S7 [6] (data from MRSA252) SACOL2225 (rpsH) 30S ribosomal protein S8 [6] (data from MRSA252) SACOL2215 (rpsM) 30S ribosomal protein S13 [6] (data from MRSA252) SACOL1292 (rpsO) 30S ribosomal protein S15 [6] (data from MRSA252) SACOL2230 (rpsQ) 30S ribosomal protein S17 [6] (data from MRSA252) SACOL2235 (rpsS) 30S ribosomal protein S19 [6] (data from MRSA252) SACOL0095 (spa) immunoglobulin G binding protein A precursor [6] (data from MRSA252) SACOL1448 (sucB) dihydrolipoamide succinyltransferase [6] (data from MRSA252) SACOL1262 (sucC) succinyl-CoA synthetase subunit beta [6] (data from MRSA252) SACOL1729 (thrS) threonyl-tRNA synthetase [6] (data from MRSA252) SACOL1722 (tig) trigger factor [6] (data from MRSA252) SACOL1377 (tkt) transketolase [6] (data from MRSA252) SACOL1276 (tsf) elongation factor Ts [6] (data from MRSA252) SACOL0594 (tuf) elongation factor Tu [6] (data from MRSA252) SACOL0211 acetyl-CoA acetyltransferase [6] (data from MRSA252) SACOL0564 pyridoxal biosynthesis lyase PdxS [6] (data from MRSA252) SACOL0731 LysR family transcriptional regulator [6] (data from MRSA252) SACOL0815 ribosomal subunit interface protein [6] (data from MRSA252) SACOL0944 NADH dehydrogenase [6] (data from MRSA252) SACOL1753 universal stress protein [6] (data from MRSA252) SACOL1759 universal stress protein [6] (data from MRSA252) SACOL1952 ferritins family protein [6] (data from MRSA252) SACOL2173 alkaline shock protein 23 [6] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: data available for NCTC8325
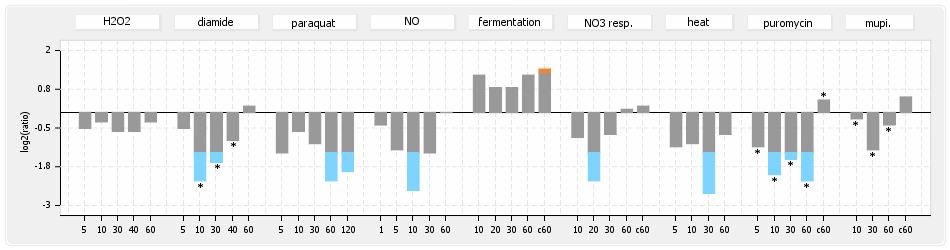
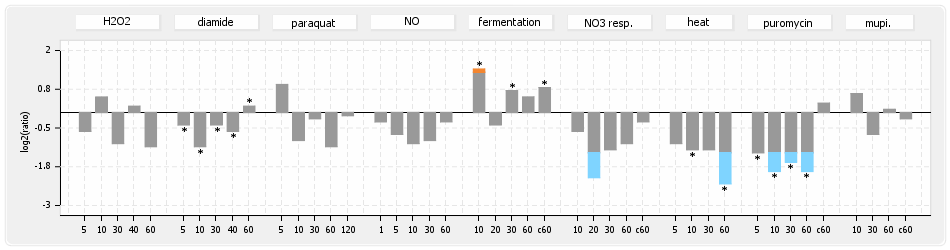
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Jan Pané-Farré, Andreas Otto, Susanne Sievers, Michael Hecker, Dörte Becher
Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach.
J Proteome Res: 2010, 9(3);1579-90
[PubMed:20108986] [WorldCat.org] [DOI] (I p) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 6.12 6.13 6.14 6.15 6.16 6.17 6.18 6.19 6.20 6.21 6.22 6.23 6.24 6.25 6.26 6.27 6.28 6.29 6.30 6.31 6.32 6.33 6.34 6.35 6.36 6.37 6.38 6.39 6.40 6.41 6.42 6.43 6.44 6.45 6.46 6.47 6.48 6.49 6.50 6.51 6.52 6.53 6.54 6.55 6.56 6.57 6.58 6.59 6.60 6.61 6.62 6.63 6.64 6.65 6.66 6.67 6.68 6.69 6.70 6.71 6.72 6.73 6.74 6.75 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p)