Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02023
- pan locus tag?: SAUPAN001702000
- symbol: SAOUHSC_02023
- pan gene symbol?: —
- synonym:
- product: bifunctional autolysin
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02023
- symbol: SAOUHSC_02023
- product: bifunctional autolysin
- replicon: chromosome
- strand: -
- coordinates: 1928152..1930050
- length: 1899
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3920477 NCBI
- RefSeq: YP_500520 NCBI
- BioCyc: G1I0R-1917 BioCyc
- MicrobesOnline: 1290476 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861ATGGGATTACCTAATCCGAAAAATAGAAAGCCCACAGCTAGTGAAGTGGTTGAATGGGCG
TTATATATCGCTAAAAACAAAATAGCTATTGATGTACCTGGTTCTGGAATGGGAGCACAA
TGCTGGGATTTACCTAATTATTTACTCGATAAATATTGGGGGTTTAGAACATGGGGAAAT
GCTGATGCTATGGCTCAAAAATCCAATTATAGAGGTAGAGATTTCAAGATAATTAGAAAT
ACAAAAGATTTTGTACCACAACCAGGCGACTGGGGTGTTTGGACTGGTGGTTGGGCAGGA
CATGTAAACATTGTAGTGGGACCATGCACAAAAGACTATTGGTATGGCGTAGATCAAAAC
TGGTATACAAATAACGCAACAGGAAGTCCACCTTATAAAATTAAACACTCTTATCATGAT
GGACCAGGTGGAGGGGTTAAATATTTTGTTAGACCACCATATCATCCAGACAAAACTACA
CCGGCACCTAAACCAGAAGATGATAGTGATGATAACGAAAAAAATAATAAAAAAGTTCCA
ATTTGGAAAGATGTAACAACTATAAAGTACACTATTTCTAGCCAAGAGGTTAATTATCCA
GAATATATTTATCACTTTATAGTAGAAGGTAATCGACGACTCGAAAAACCTAAAGGAATA
ATGATTAGAAACGCACAAACGATGAGCTCGGTAGAAAGTTTATATAACAGTAGGAAGAAA
TACAAACAGGATGTAGAATATCCCCACTTTTATGTTGATAGACATAATATTTGGGCACCT
AGAAGAGCTGTATTTGAAGTTCCTAATGAACCTGATTATATAGTTATAGACGTATGTGAA
GATTATAGTGCGAGTAAAAATGAATTTATTTTTAATGAGATTCACGCAATGGTTGTAGCT
GTAGATATGATGGCCAAATATGAGATACCTCTAAGTATTGAAAATTTAAAAGTAGACGAC
AGCATTTGGCGTTCAATGTTGGAACATGTTAATTGGAATATGATTGACAACGGTGTTCCT
CCTAAAGATAAATACGAAGCATTAGAAAAGGCATTACTTAATATATTTAAAAACAGAGAA
AAATTATTAAATTCTATAACTAAGCCAACAGTAACAAAATCTAGAATAAAAGTTATGGTA
GATAATAAAAACGCTGATATAGCTAATGTAAGAGACTCGTCACCAACAGCCAACAATGGT
TCGGCATCTAAACAACCGCAGATCATAACAGAAACGAGTCCTTATACATTCAAACAAGCA
CTGGATAAACAAATGGCAAGAGGTAACCCGAAAAAATCTAATGCTTGGGGTTGGGCTAAC
GCTACACGAGCACAAACGAGTTCAGCAATGAATGTAAAGCGTATATGGGAAAGTAACACA
CAATGCTACCAAATGCTTAATTTAGGCAAGTATCAAGGTGTTTCAGTTAGCGCACTTAAT
AAGATACTTAAAGGTAAGGGAACATTGAATAATCAAGGTAAAGCGTTCGCAGAAGCTTGT
AAAAAGCACAACATTAATGAAATTTATTTAATCGCGCATGCTTTCTTAGAAAGTGGATAT
GGAACAAGTAACTTCGCTAACGGAAAAGATGGAGTATACAACTACTTCGGCATTGGCGCT
TACGACAACAATCCTAACTACGCAATGACGTTTGCAAGGAATAAAGGTTGGACATCTCCA
GCAAAAGCAATCATGGGCGGTGCTAGCTTCGTAAGAAAGGATTACATCAATAAAGGTCAA
AACACATTGTACCGAATTAGATGGAATCCTAAGAATCCAGCTACCCACCAATACGCTACT
GCTATAGAGTGGTGCCAACATCAAGCAAGTACAATCGCTAAGTTATATAAACAAATCGGC
TTAAAAGGTATCTACTTCACAAGGGATAAATATAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1899
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02023
- symbol: SAOUHSC_02023
- description: bifunctional autolysin
- length: 632
- theoretical pI: 9.77316
- theoretical MW: 72002.1
- GRAVY: -0.688608
⊟Function[edit | edit source]
- reaction: EC 3.5.1.28? ExPASyN-acetylmuramoyl-L-alanine amidase Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides
- TIGRFAM: flagellar rod assembly protein/muramidase FlgJ (TIGR02541; HMM-score: 22.1)
- TheSEED :
- Phage endo-beta-N-acetylglucosaminidase (EC 3.2.1.96)
- Phage endolysin N-acetylmuramoyl-L-alanine amidase (EC 3.5.1.28)
Cell Wall and Capsule Cell Wall and Capsule - no subcategory Murein Hydrolases N-acetylmuramoyl-L-alanine amidase (EC 3.5.1.28)and 3 moreCell Wall and Capsule Cell Wall and Capsule - no subcategory Recycling of Peptidoglycan Amino Acids N-acetylmuramoyl-L-alanine amidase (EC 3.5.1.28) - PFAM: Lysozyme (CL0037) Glucosaminidase; Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase (PF01832; HMM-score: 51.4)and 2 moreno clan defined DUF5415; Family of unknown function (DUF5415) (PF17436; HMM-score: 13.8)Lysozyme (CL0037) SLT_2; Transglycosylase SLT domain (PF13406; HMM-score: 13.4)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Extracellular
- Cytoplasmic Score: 0.01
- Cytoplasmic Membrane Score: 0.09
- Cellwall Score: 0.18
- Extracellular Score: 9.72
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0.0001
- Cytoplasmic Membrane Score: 0
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.9998
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.031405
- TAT(Tat/SPI): 0.001237
- LIPO(Sec/SPII): 0.002792
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MGLPNPKNRKPTASEVVEWALYIAKNKIAIDVPGSGMGAQCWDLPNYLLDKYWGFRTWGNADAMAQKSNYRGRDFKIIRNTKDFVPQPGDWGVWTGGWAGHVNIVVGPCTKDYWYGVDQNWYTNNATGSPPYKIKHSYHDGPGGGVKYFVRPPYHPDKTTPAPKPEDDSDDNEKNNKKVPIWKDVTTIKYTISSQEVNYPEYIYHFIVEGNRRLEKPKGIMIRNAQTMSSVESLYNSRKKYKQDVEYPHFYVDRHNIWAPRRAVFEVPNEPDYIVIDVCEDYSASKNEFIFNEIHAMVVAVDMMAKYEIPLSIENLKVDDSIWRSMLEHVNWNMIDNGVPPKDKYEALEKALLNIFKNREKLLNSITKPTVTKSRIKVMVDNKNADIANVRDSSPTANNGSASKQPQIITETSPYTFKQALDKQMARGNPKKSNAWGWANATRAQTSSAMNVKRIWESNTQCYQMLNLGKYQGVSVSALNKILKGKGTLNNQGKAFAEACKKHNINEIYLIAHAFLESGYGTSNFANGKDGVYNYFGIGAYDNNPNYAMTFARNKGWTSPAKAIMGGASFVRKDYINKGQNTLYRIRWNPKNPATHQYATAIEWCQHQASTIAKLYKQIGLKGIYFTRDKYK
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_02022 < SAOUHSC_02023predicted SigA promoter [1] : S810 < SAOUHSC_02019 < SAOUHSC_02020 < SAOUHSC_02021 < SAOUHSC_02022 < SAOUHSC_02023 < SAOUHSC_02025 < SAOUHSC_02026 < SAOUHSC_02027 < SAOUHSC_02028 < SAOUHSC_02029 < SAOUHSC_02030 < SAOUHSC_02031 < SAOUHSC_02033 < SAOUHSC_02034 < SAOUHSC_02035 < SAOUHSC_02036 < SAOUHSC_02037 < SAOUHSC_02038 < SAOUHSC_02040 < SAOUHSC_02041 < SAOUHSC_02042 < SAOUHSC_02043 < SAOUHSC_02044 < S811 < SAOUHSC_02046 < SAOUHSC_02047 < SAOUHSC_02048 < SAOUHSC_02049 < SAOUHSC_02050 < S812 < SAOUHSC_02051 < SAOUHSC_02052 < SAOUHSC_02053 < SAOUHSC_02054 < SAOUHSC_02055 < SAOUHSC_02056 < SAOUHSC_02057 < SAOUHSC_02058 < SAOUHSC_02059 < SAOUHSC_02060 < SAOUHSC_02061 < SAOUHSC_02062 < SAOUHSC_02063 < SAOUHSC_02064 < SAOUHSC_02065 < SAOUHSC_02066 < SAOUHSC_02067 < SAOUHSC_02068 < SAOUHSC_02069 < SAOUHSC_02070 < SAOUHSC_02071 < S813 < SAOUHSC_02072 < SAOUHSC_02073 < SAOUHSC_02074 < SAOUHSC_02075 < SAOUHSC_02076 < SAOUHSC_02077 < SAOUHSC_02078 < SAOUHSC_02079 < SAOUHSC_02080 < S816 < SAOUHSC_02081 < SAOUHSC_02083
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
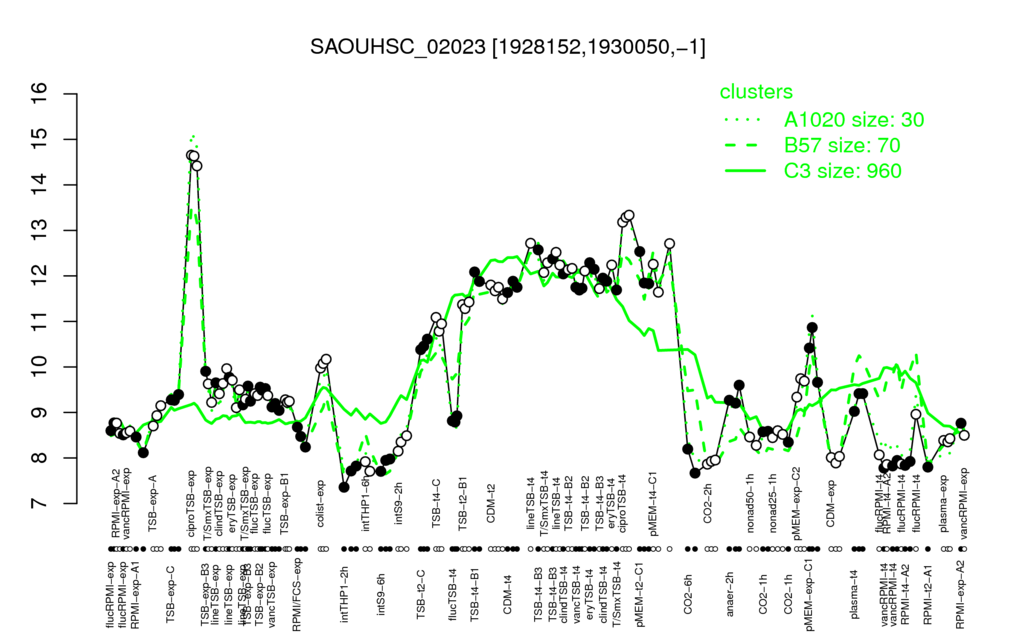 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
