Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02030
- pan locus tag?: SAUPAN001697000
- symbol: SAOUHSC_02030
- pan gene symbol?: —
- synonym:
- product: phi ETA orf 55-like protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02030
- symbol: SAOUHSC_02030
- product: phi ETA orf 55-like protein
- replicon: chromosome
- strand: -
- coordinates: 1934828..1936729
- length: 1902
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3920483 NCBI
- RefSeq: YP_500526 NCBI
- BioCyc: G1I0R-1923 BioCyc
- MicrobesOnline: 1290482 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861ATGACAATAACTATTAAACCACCTAAAGGTAATGGCGCACCTGTACCAGTAGAAACAACT
TTAGTAAAAAAAGTTAATGCTGACGGTGTATTAACTTTTGATATTCTAGAAAATAAATAT
ACTTATGAAGTTATTAACGCTATAGGGAAAAGATGGATTGTTAGTCATGTCGAAGGTGAA
AACGACAAGAAAGAATATGTAATAACTGTCATTGATAGGAAATCAGAAGGCGACAGACAA
CTGGTTGAATGTACTGCTAGAGAGATTCCCATAGACAAGTTAATGATTGATAGAATTTAT
GTTAATGTAACAGGATCTTTTACAGTAGAAAGATATTTTAACATTGTGTTTCAAGGTACT
GGAATGCTTTTTGAAGTCGAGGGCAAAGTTAAATCTTCAAAGTTTGAAAATGGTGGTGAA
GGCGATACAAGGTTAGAAATGTTTAAAAAGGGATTAGAACATTTCGGTTTAGAATATAAA
ATAACGTATGACAAAAAGAAAGACAGATATAAGTTTGTATTGACGCCTTTTGCAAATCAA
AAAGCGTCTTATTTTATTTCTGACGAAGTCAACGCCAACGCTATAAAACTCGAGGAAGAT
GCAAGTGATTTCGCCACCTTCATTAGAGGATATGGTAATTATTCAGGAGAAGAAACATTC
GAACACGCTGGGCTCGTAATGGAAGCTAGAAGTGCATTAGCTGAAATATACGGCGACATC
CACGCAGAACCATTTAAAGATGGTAAAGTGACTGACCAAGAAACTATGGATAAAGAATTA
CAATCGAGATTGAAAAAGTCGTTAAAACAATCTTTGTCTTTGGACTTTTTGGTGTTAAGA
GAATCATATCCAGAAGCAGACCCACAACCCGGAGACATAGTACAAATAAAATCTACCAAA
CTAGGTTTGAATGATTTAGTCCGTATAGTACAAGTTAAAACGATTAGGGGTATAAACAAT
GTAATTGTTAAGCAAGATGTAACGCTTGGTGAGTTTAATCGAGAACAACGATATATGAAA
AAAGTTAATACTGCAGCTAACTATGTTTCTGGATTAAATGATGTTAACCTTTCTAATCCT
AGTAAAGCGGCAGAAAACTTGAAGTCTAAAGTAGCGTCAATAGCTAAATCAACACTCGAT
TTGATGAGTAGAACTGATTTGATTGAAGATAAACAACAGAAGGTAAGCTCTAAAACTGTG
ACTACATCTGACGGCACTATCGTTCATGATTTTATAGATAAATCAAACATTAAAGATGTA
AAAACGATTGGAACGATTGGCGATTCTGTAGCTAGAGGATCACATGCGAAAACTAATTTC
ACAGAAATGTTAGGCAAGAAGTTAAAAGCTAAAACGACCAACCTTGCAAGAGGTGGCGCA
ACAATGGCAACAGTTCCAATAGGTAAAGAAGCGGTAGAAAACAGCATTTATAGACAAGCA
GAGCAAATAAGAGGAGACCTAATCATATTACAAGGTACAGATGATGACTGGTTACATGGT
TATTGGGCAGGCGTACCGATAGGCACTGATAAAACCGACACTAAAACGTTTTACGGCGCC
TTTTGTTCTGCAATTGAAGTTATCAGGAAAAATAATCCAGCTTCAAAAATACTTGTAATG
ACAGCTACTAGGCAATGCCCTATGAGTGGTACAACGATACGCCGTAAAGATACGGACAAA
AACAAACTAGGGTTAACTTTAGAGGATTATGTCAATGCTCAGATATTGGCTTGTAGTGAA
TTGGATGTACCAGTATATGATGCTTATCACACAGATTATTTCAAACCATATAATCCAGCA
TTTAGAAAATCTAGCATGCCTGATGGATTACATCCTAATGAAAGAGGTCATGAAGTTATT
ATGTATGAGCTTATTAAAAATTATTATCAGTTTTATGGATAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1902
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02030
- symbol: SAOUHSC_02030
- description: phi ETA orf 55-like protein
- length: 633
- theoretical pI: 7.3187
- theoretical MW: 70987.2
- GRAVY: -0.452765
⊟Function[edit | edit source]
- TIGRFAM:
- TheSEED :
- Phage phi 11 orf44 protein homolog
- PFAM: no clan defined TT1_Tal; Tal, N-terminal tail tube TT1 domain (PF24650; HMM-score: 116.6)and 4 morePhage_barrel (CL0504) Prophage_tail; Prophage endopeptidase tail (PF06605; HMM-score: 79.6)SGNH_hydrolase (CL0264) Lipase_GDSL_2; GDSL-like Lipase/Acylhydrolase family (PF13472; HMM-score: 55.1)Lipase_GDSL; GDSL-like Lipase/Acylhydrolase (PF00657; HMM-score: 40.2)no clan defined DUF5839; Family of unknown function (DUF5839) (PF19157; HMM-score: 12.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0.0698
- Cytoplasmic Membrane Score: 0.0231
- Cell wall & surface Score: 0.0998
- Extracellular Score: 0.8073
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 0
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.008885
- TAT(Tat/SPI): 0.000504
- LIPO(Sec/SPII): 0.000846
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTITIKPPKGNGAPVPVETTLVKKVNADGVLTFDILENKYTYEVINAIGKRWIVSHVEGENDKKEYVITVIDRKSEGDRQLVECTAREIPIDKLMIDRIYVNVTGSFTVERYFNIVFQGTGMLFEVEGKVKSSKFENGGEGDTRLEMFKKGLEHFGLEYKITYDKKKDRYKFVLTPFANQKASYFISDEVNANAIKLEEDASDFATFIRGYGNYSGEETFEHAGLVMEARSALAEIYGDIHAEPFKDGKVTDQETMDKELQSRLKKSLKQSLSLDFLVLRESYPEADPQPGDIVQIKSTKLGLNDLVRIVQVKTIRGINNVIVKQDVTLGEFNREQRYMKKVNTAANYVSGLNDVNLSNPSKAAENLKSKVASIAKSTLDLMSRTDLIEDKQQKVSSKTVTTSDGTIVHDFIDKSNIKDVKTIGTIGDSVARGSHAKTNFTEMLGKKLKAKTTNLARGGATMATVPIGKEAVENSIYRQAEQIRGDLIILQGTDDDWLHGYWAGVPIGTDKTDTKTFYGAFCSAIEVIRKNNPASKILVMTATRQCPMSGTTIRRKDTDKNKLGLTLEDYVNAQILACSELDVPVYDAYHTDYFKPYNPAFRKSSMPDGLHPNERGHEVIMYELIKNYYQFYG
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_02028 < SAOUHSC_02029 < SAOUHSC_02030 < SAOUHSC_02031predicted SigA promoter [1] : S810 < SAOUHSC_02019 < SAOUHSC_02020 < SAOUHSC_02021 < SAOUHSC_02022 < SAOUHSC_02023 < SAOUHSC_02025 < SAOUHSC_02026 < SAOUHSC_02027 < SAOUHSC_02028 < SAOUHSC_02029 < SAOUHSC_02030 < SAOUHSC_02031 < SAOUHSC_02033 < SAOUHSC_02034 < SAOUHSC_02035 < SAOUHSC_02036 < SAOUHSC_02037 < SAOUHSC_02038 < SAOUHSC_02040 < SAOUHSC_02041 < SAOUHSC_02042 < SAOUHSC_02043 < SAOUHSC_02044 < S811 < SAOUHSC_02046 < SAOUHSC_02047 < SAOUHSC_02048 < SAOUHSC_02049 < SAOUHSC_02050 < S812 < SAOUHSC_02051 < SAOUHSC_02052 < SAOUHSC_02053 < SAOUHSC_02054 < SAOUHSC_02055 < SAOUHSC_02056 < SAOUHSC_02057 < SAOUHSC_02058 < SAOUHSC_02059 < SAOUHSC_02060 < SAOUHSC_02061 < SAOUHSC_02062 < SAOUHSC_02063 < SAOUHSC_02064 < SAOUHSC_02065 < SAOUHSC_02066 < SAOUHSC_02067 < SAOUHSC_02068 < SAOUHSC_02069 < SAOUHSC_02070 < SAOUHSC_02071 < S813 < SAOUHSC_02072 < SAOUHSC_02073 < SAOUHSC_02074 < SAOUHSC_02075 < SAOUHSC_02076 < SAOUHSC_02077 < SAOUHSC_02078 < SAOUHSC_02079 < SAOUHSC_02080 < S816 < SAOUHSC_02081 < SAOUHSC_02083
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
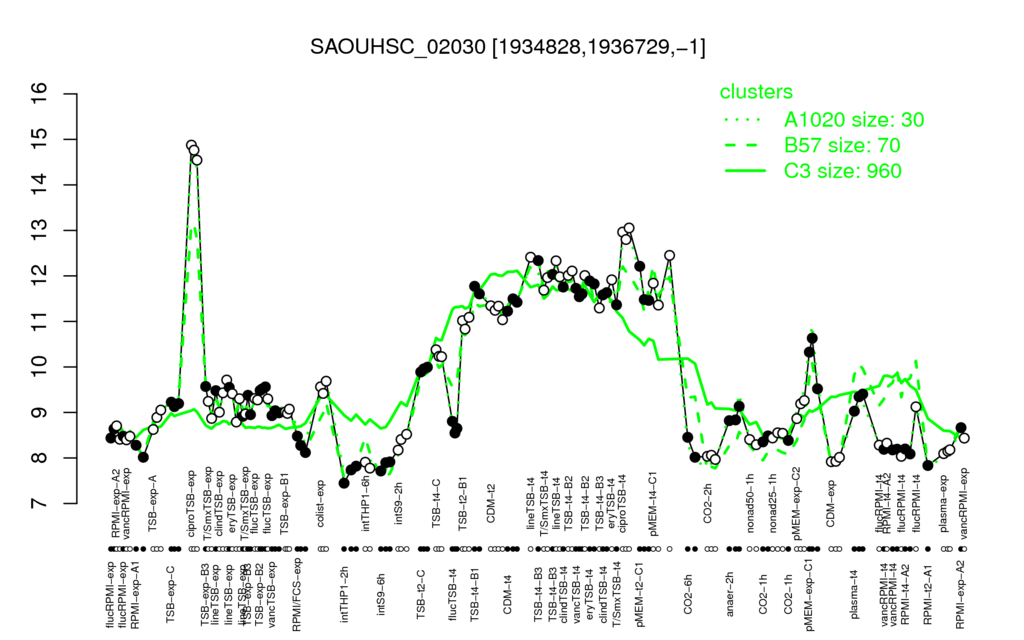 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
