Jump to navigation
Jump to search
COLN315NCTC8325NewmanUSA300_FPR375704-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159JSNZLGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02029
- pan locus tag?:
- symbol: SAOUHSC_02029
- pan gene symbol?: —
- synonym:
- product: phi ETA orf 56-like protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02029
- symbol: SAOUHSC_02029
- product: phi ETA orf 56-like protein
- replicon: chromosome
- strand: -
- coordinates: 1932903..1934813
- length: 1911
- essential: unknown
⊟Accession numbers[edit | edit source]
- Gene ID: 3920482 NCBI
- RefSeq: YP_500525 NCBI
- BioCyc: G1I0R-1922 BioCyc
- MicrobesOnline: 1290481 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861ATGAGTAATAAACTAATTACAGATTTAAGTAGAGTCTTTGACTACAGATATGTAGATGAA
AATGAGTATAACTTTAAACTTATTTCAGACATGCTGACGGATTTTAATTTCTCTCTTGAA
TACCACAGAAATAAAGAGGTATTCGCACATGATGGAGAACAAATAAAGTATGAACATTTA
AATGTTACAAGTAACGTCTCTGACTTTTTAACATATTTAAACGGTCGATTTAGCAACATG
GTACTAGGTCATAACGGCGACGGTATCAACGAAGTAAAAGACGCGCGCGTTGATAATACA
GGTTATGGTCATAAGACATTGCAAGATCGTTTGTATCATGATTATTCAACACTAGATGTT
TTCACTAAAAAGGTTGAGAAAGCTGTAGATGAACACTATAAAGAATATCGAGCGACAGAA
TACCGATTCGAACCAAAAGAGCAAGAACCGGAATTTATCACTGATTTATCGCCATATACA
AATGCAGTAATGCAATCATTTTGGGTAGACCCTAGAACGAAAATTATTTATATGACGCAA
GCTCGTCCAGGTAATCATTACATGTTATCTAGATTGAAGCCCAACGGACAATTTATTGAT
AGATTGCTTGTTAAAAACGGCGGTCACGGTACACACAATGCGTATAGATACATTGATGGA
GAATTATGGATTTATTCAGCTGTATTGGACAGTAACAAAAACAACAAGTTTGTACGTTTC
CAATATAGAACTGGAGAAATAACTTATGGTAATGAAATGCAAGATGTCATGCCGAATATA
TTTAACGACAGATATACGTCAGCGATTTATAATCCGGTAGAAAATTTAATGATTTTTAGA
CGTGAATATAAACCCACTGAAAGACAACTTAAGAATTCGTTGAACTTTGTTGAGGTTAGA
AGTGCTGACGATATTGATAAAGGTATAGACAAAGTATTGTATCAAATGGATATACCTATG
GAATACACTTCAGATACACAACCTATGCAAGGTATCACTTATGATGCAGGTATCTTATAT
TGGTATACAGGTGATTCGAATACAGCCAACCCTAACTACTTACAAGGCTTCGATATCAAA
ACGAAAGAATTGTTATTTAAACGTCGCATCGATATAGGCGGTGTGAATAACAACTTTAAA
GGAGATTTCCAAGAGGCTGAGGGTCTAGATATGTATTACGATCTAGAAACAGGACGTAAA
GCACTTCTAATCGGGGTAACTATTGGACCTGGTAACAACAGACATCATTCAATTTATTCT
ATCGGTCAAAGAGGTGTAAACCAATTCTTGAAAAACATCGCACCTCAAGTATCAATGACT
GATTCAGGCGGACGTGTTAAACCGTTACCAATACAGAACCCAGCATATCTAAGTGATATT
ACGGAAGTTGGTCATTACTATATCTATACGCAAGACACACAAAATGCATTAGATTTCCCG
TTACCGAAAGCGTTTAGAGATGCAGGGTGGTTCTTGGATGTACTGCCTGGACACTATAAT
GGTGCTCTAAGACAAGTACTTACCAGAAACAGCACAGGTAGAAATATGCTTAAATTCGAA
CGTGTCATTGACATTTTCAATAAGAAAAACAACGGAGCATGGAATTTCTGCCCGCAAAAC
GCCGGTTATTGGGAACATATCCCTAAGAGTATTACAAAATTATCAGATTTAAAAATCGTT
GGTTTAGATTTCTATATCACTACTGAAGAATCAAACCGATTTACTGATTTTCCTAAAGAC
TTTAAAGGTATTGCAGGTTGGATATTAGAAGTAAAATCGAATACACCAGGTAATACAACA
CAAGTATTAAGACGTAATAACTTCCCGTCTGCACATCAATTTTTAGTTAGAAACTTTGGT
ACTGGTGGCGTTGGTAAATGGAGTTTATTCGAAGGAAAGGTGGTTGAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1911
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02029
- symbol: SAOUHSC_02029
- description: phi ETA orf 56-like protein
- length: 636
- theoretical pI: 6.73317
- theoretical MW: 73649.2
- GRAVY: -0.622013
⊟Function[edit | edit source]
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MSNKLITDLSRVFDYRYVDENEYNFKLISDMLTDFNFSLEYHRNKEVFAHDGEQIKYEHLNVTSNVSDFLTYLNGRFSNMVLGHNGDGINEVKDARVDNTGYGHKTLQDRLYHDYSTLDVFTKKVEKAVDEHYKEYRATEYRFEPKEQEPEFITDLSPYTNAVMQSFWVDPRTKIIYMTQARPGNHYMLSRLKPNGQFIDRLLVKNGGHGTHNAYRYIDGELWIYSAVLDSNKNNKFVRFQYRTGEITYGNEMQDVMPNIFNDRYTSAIYNPVENLMIFRREYKPTERQLKNSLNFVEVRSADDIDKGIDKVLYQMDIPMEYTSDTQPMQGITYDAGILYWYTGDSNTANPNYLQGFDIKTKELLFKRRIDIGGVNNNFKGDFQEAEGLDMYYDLETGRKALLIGVTIGPGNNRHHSIYSIGQRGVNQFLKNIAPQVSMTDSGGRVKPLPIQNPAYLSDITEVGHYYIYTQDTQNALDFPLPKAFRDAGWFLDVLPGHYNGALRQVLTRNSTGRNMLKFERVIDIFNKKNNGAWNFCPQNAGYWEHIPKSITKLSDLKIVGLDFYITTEESNRFTDFPKDFKGIAGWILEVKSNTPGNTTQVLRRNNFPSAHQFLVRNFGTGGVGKWSLFEGKVVE
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_02028 < SAOUHSC_02029 < SAOUHSC_02030 < SAOUHSC_02031predicted SigA promoter [3] : S810 < SAOUHSC_02019 < SAOUHSC_02020 < SAOUHSC_02021 < SAOUHSC_02022 < SAOUHSC_02023 < SAOUHSC_02025 < SAOUHSC_02026 < SAOUHSC_02027 < SAOUHSC_02028 < SAOUHSC_02029 < SAOUHSC_02030 < SAOUHSC_02031 < SAOUHSC_02033 < SAOUHSC_02034 < SAOUHSC_02035 < SAOUHSC_02036 < SAOUHSC_02037 < SAOUHSC_02038 < SAOUHSC_02040 < SAOUHSC_02041 < SAOUHSC_02042 < SAOUHSC_02043 < SAOUHSC_02044 < S811 < SAOUHSC_02046 < SAOUHSC_02047 < SAOUHSC_02048 < SAOUHSC_02049 < SAOUHSC_02050 < S812 < SAOUHSC_02051 < SAOUHSC_02052 < SAOUHSC_02053 < SAOUHSC_02054 < SAOUHSC_02055 < SAOUHSC_02056 < SAOUHSC_02057 < SAOUHSC_02058 < SAOUHSC_02059 < SAOUHSC_02060 < SAOUHSC_02061 < SAOUHSC_02062 < SAOUHSC_02063 < SAOUHSC_02064 < SAOUHSC_02065 < SAOUHSC_02066 < SAOUHSC_02067 < SAOUHSC_02068 < SAOUHSC_02069 < SAOUHSC_02070 < SAOUHSC_02071 < S813 < SAOUHSC_02072 < SAOUHSC_02073 < SAOUHSC_02074 < SAOUHSC_02075 < SAOUHSC_02076 < SAOUHSC_02077 < SAOUHSC_02078 < SAOUHSC_02079 < SAOUHSC_02080 < S816 < SAOUHSC_02081 < SAOUHSC_02083
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
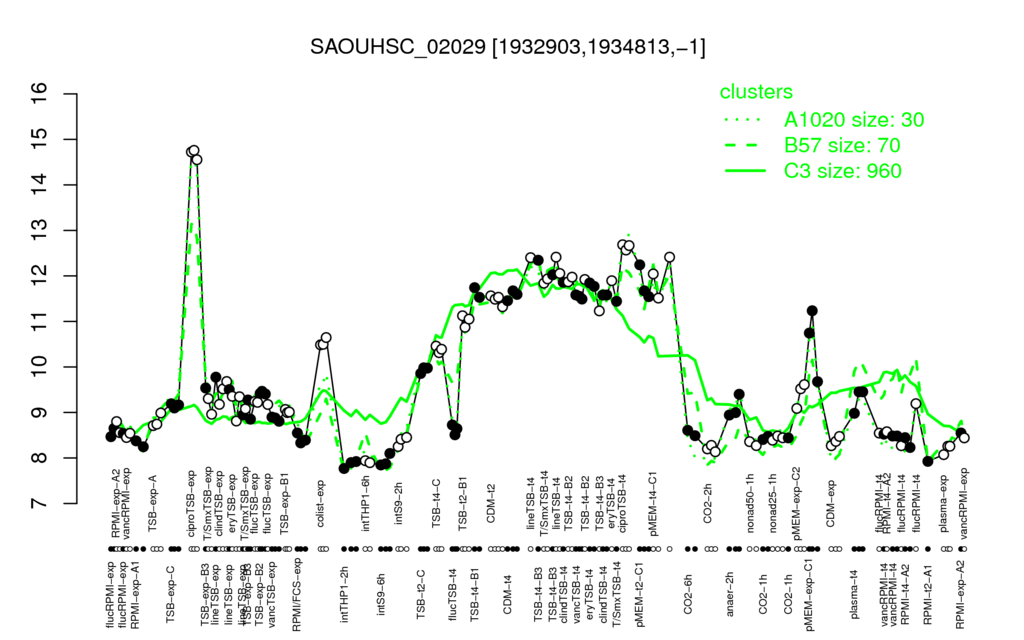 ⊟Multi-gene expression profiles
⊟Multi-gene expression profiles
Click on any data point to display a description of the corresponding condition!
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊞Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊞Other Information[edit | edit source]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Jump up to: 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
