Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02287
- pan locus tag?: SAUPAN005309000
- symbol: SAOUHSC_02287
- pan gene symbol?: leuC
- synonym:
- product: isopropylmalate isomerase large subunit
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02287
- symbol: SAOUHSC_02287
- product: isopropylmalate isomerase large subunit
- replicon: chromosome
- strand: +
- coordinates: 2119308..2120678
- length: 1371
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919162 NCBI
- RefSeq: YP_500769 NCBI
- BioCyc: G1I0R-2159 BioCyc
- MicrobesOnline: 1290725 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321ATGGGTCAAACATTATTTGACAAGGTGTGGAACAGACATGTGTTATACGGGAAATTGGGC
GAACCGCAACTATTATACATTGATTTACACCTTATACATGAAGTTACTTCTCCTCAAGCA
TTTGAAGGACTTAGGCTTCAAAACAGAAAATTAAGACGCCCAGATTTAACATTTGCAACA
CTCGATCACAATGTTCCTACTATTGATATATTCAATATTAAAGATGAAATTGCAAACAAA
CAAATCACAACATTACAAAAAAACGCCATAGATTTTGGGGTGCATATTTTTGATATGGGT
TCTGATGAACAAGGTATTGTTCACATGGTAGGACCTGAGACAGGACTTACACAGCCTGGC
AAGACAATCGTTTGTGGTGACTCTCACACAGCAACACATGGAGCATTTGGTGCTATTGCA
TTTGGAATTGGAACAAGTGAAGTTGAACATGTTTTCGCAACTCAAACGCTATGGCAAACA
AAACCCAAAAACTTAAAAATCGATATTAATGGTACCTTACCAACAGGCGTCTATGCTAAG
GACATTATTCTGCATTTAATTAAAACGTATGGTGTTGACTTTGGTACAGGCTATGCTTTG
GAATTTACTGGCGAAACAATTAAAAACCTTTCAATGGATGGTCGAATGACTATTTGTAAC
ATGGCTATCGAAGGTGGTGCCAAATACGGCATAATCCAACCTGATGATATAACATTTGAA
TATGTTAAAGGGAGACCATTTGCCGATAACTTCGCTAAATCAGTTGATAAGTGGCGTGAG
CTATATTCTGATGACGACGCGATATTTGATCGTGTAATTGAACTTGATGTTTCAACATTA
GAACCACAAGTGACATGGGGAACTAATCCTGAAATGGGTGTTAATTTCAGTGAACCATTC
CCTGAAATCAATGATATCAACGATCAACGTGCGTATGATTATATGGGGTTAGAACCAGGT
CAAAAAGCTGAAGACATCGACTTAGGGTATGTTTTTCTCGGTTCATGTACAAATGCTAGA
CTATCAGATTTGATTGAAGCTAGTCATATTGTTAAAGGAAATAAAGTTCATCCAAATATT
ACAGCTATTGTCGTACCAGGTTCTCGTACAGTAAAAAAAGAAGCAGAAAAATTAGGTCTA
GATACTATCTTTAAAAATGCAGGATTTGAATGGCGTGAACCAGGATGTTCAATGTGTTTA
GGCATGAATCCTGACCAAGTACCTGAGGGCGTACATTGTGCATCTACAAGTAATCGAAAC
TTTGAAGGACGACAAGGCAAAGGTGCAAGAACACATTTAGTATCCCCTGCTATGGCAGCA
GCAGCAGCTATTCATGGTAAATTTGTGGACGTAAGAAAGGTGGTTGTTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1371
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02287
- symbol: SAOUHSC_02287
- description: isopropylmalate isomerase large subunit
- length: 456
- theoretical pI: 5.56699
- theoretical MW: 50325.9
- GRAVY: -0.230263
⊟Function[edit | edit source]
- ⊞reaction: EC 4.2.1.33? ExPASy
- ⊞TIGRFAM: Amino acid biosynthesis Pyruvate family 3-isopropylmalate dehydratase, large subunit (TIGR00170; EC 4.2.1.33; HMM-score: 695.6)and 9 more
- ⊞TheSEED :
- 3-isopropylmalate dehydratase large subunit (EC 4.2.1.33)
Amino Acids and Derivatives Branched-chain amino acids Branched-Chain Amino Acid Biosynthesis 3-isopropylmalate dehydratase large subunit (EC 4.2.1.33)and 1 more - ⊞PFAM: no clan defined Aconitase; Aconitase family (aconitate hydratase) (PF00330; HMM-score: 560.8)and 1 more
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: [4Fe-4S] cluster
- effectors:
⊟Localization[edit | edit source]
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MGQTLFDKVWNRHVLYGKLGEPQLLYIDLHLIHEVTSPQAFEGLRLQNRKLRRPDLTFATLDHNVPTIDIFNIKDEIANKQITTLQKNAIDFGVHIFDMGSDEQGIVHMVGPETGLTQPGKTIVCGDSHTATHGAFGAIAFGIGTSEVEHVFATQTLWQTKPKNLKIDINGTLPTGVYAKDIILHLIKTYGVDFGTGYALEFTGETIKNLSMDGRMTICNMAIEGGAKYGIIQPDDITFEYVKGRPFADNFAKSVDKWRELYSDDDAIFDRVIELDVSTLEPQVTWGTNPEMGVNFSEPFPEINDINDQRAYDYMGLEPGQKAEDIDLGYVFLGSCTNARLSDLIEASHIVKGNKVHPNITAIVVPGSRTVKKEAEKLGLDTIFKNAGFEWREPGCSMCLGMNPDQVPEGVHCASTSNRNFEGRQGKGARTHLVSPAMAAAAAIHGKFVDVRKVVV
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [1] : S883 > S884 > SAOUHSC_02281 > SAOUHSC_02282 > SAOUHSC_02283 > S885 > SAOUHSC_02284 > SAOUHSC_02285 > SAOUHSC_02286 > SAOUHSC_02287 > leuD > SAOUHSC_02289
⊟Regulation[edit | edit source]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
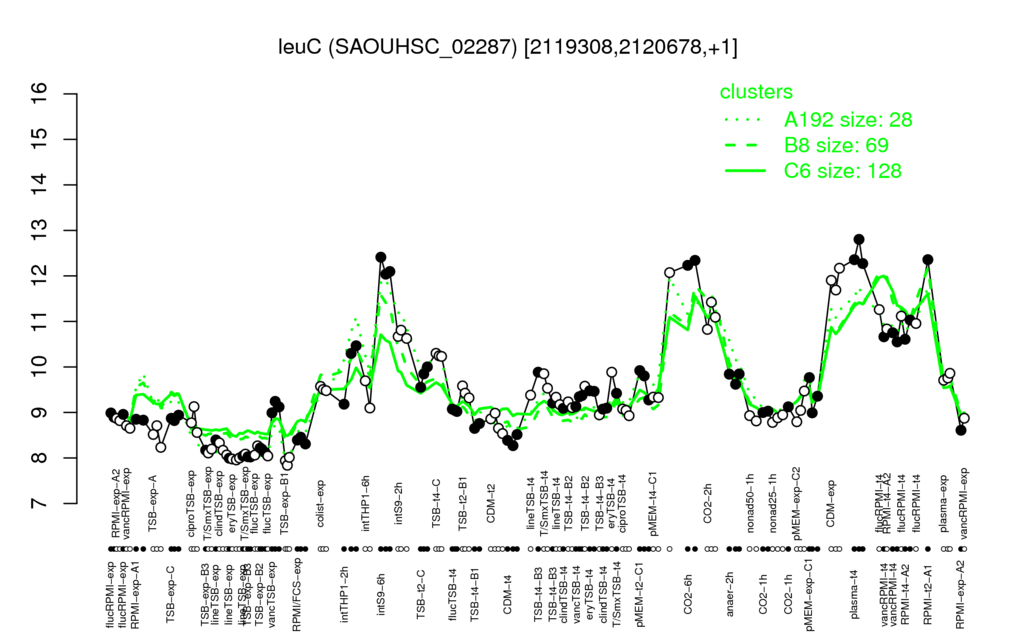 ⊟Multi-gene expression profiles
⊟Multi-gene expression profiles
Click on any data point to display a description of the corresponding condition!
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊞Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊞Other Information[edit | edit source]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Jump up to: 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ Charlotte D Majerczyk, Paul M Dunman, Thanh T Luong, Chia Y Lee, Marat R Sadykov, Greg A Somerville, Kip Bodi, Abraham L Sonenshein
Direct targets of CodY in Staphylococcus aureus.
J Bacteriol: 2010, 192(11);2861-77
[PubMed:20363936] [WorldCat.org] [DOI] (I p)
