Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01793
- pan locus tag?: SAUPAN004308000
- symbol: nrdR
- pan gene symbol?: nrdR
- synonym:
- product: transcriptional regulator NrdR
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01793
- symbol: nrdR
- product: transcriptional regulator NrdR
- replicon: chromosome
- strand: -
- coordinates: 1690427..1690897
- length: 471
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920434 NCBI
- RefSeq: YP_500297 NCBI
- BioCyc: G1I0R-1666 BioCyc
- MicrobesOnline: 1290211 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421ATGAAATGCCCGAAATGTAATTCTACACAATCTAAAGTTGTAGATTCAAGGCATGCCGAT
GAATTAAATGCCATTCGAAGACGAAGAGAATGTGAAAATTGTGGAACACGTTTCACTACA
TTTGAACATATCGAAGTTAGTCAGCTTATAGTTGTGAAAAAAGATGGCACAAGAGAGCAG
TTTTCAAGAGAAAAGATACTTAATGGACTTGTGCGTTCTTGTGAGAAACGACCAGTTAGA
TATCAACAACTTGAAGACATAACTAACAAGGTTGAATGGCAATTACGAGATGAAGGTCAT
ACGGAAGTGTCTTCACGAGATATAGGTGAACACGTTATGAACTTGTTAATGCATGTTGAT
CAAGTTTCATATGTTAGATTTGCATCAGTCTATAAAGAATTTAAAGATGTTGATCAATTG
TTAGCATCGATGCAAGGGATTTTAAGCGAAAACAAACGGAGTGATGCTTAA60
120
180
240
300
360
420
471
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01793
- symbol: NrdR
- description: transcriptional regulator NrdR
- length: 156
- theoretical pI: 7.89688
- theoretical MW: 18203.5
- GRAVY: -0.744872
⊟Function[edit | edit source]
- TIGRFAM: Regulatory functions DNA interactions transcriptional regulator NrdR (TIGR00244; HMM-score: 187.7)and 4 moreCys-rich peptide, TIGR04165 family (TIGR04165; HMM-score: 16.4)Regulatory functions DNA interactions putative regulatory protein, FmdB family (TIGR02605; HMM-score: 14.6)MJ0042 family finger-like domain (TIGR02098; HMM-score: 12)cxxc_20_cxxc protein (TIGR04104; HMM-score: 8.7)
- TheSEED :
- Ribonucleotide reductase transcriptional regulator NrdR
- PFAM: no clan defined ATP-cone; ATP cone domain (PF03477; HMM-score: 77.7)Zn_Beta_Ribbon (CL0167) Zn_ribbon_NrdR; Transcriptional repressor NrdR-like, N-terminal domain (PF22811; HMM-score: 72.3)and 18 moreC2H2-zf (CL0361) zf-DBF; DBF zinc finger (PF07535; HMM-score: 19.5)Zn_Beta_Ribbon (CL0167) Zn_Ribbon_TF; TFIIB zinc-binding (PF08271; HMM-score: 16.3)no clan defined DUF7117; Family of unknown function (DUF7117) (PF23430; HMM-score: 15.4)Zn_Beta_Ribbon (CL0167) Zn_ribbon_IS1595; Transposase zinc-ribbon domain (PF12760; HMM-score: 15)Zn_ribbon_GRF_2; GRF-like zinc ribbon domain (PF23549; HMM-score: 14.7)Zn_ribbon_8; Zinc ribbon domain (PF09723; HMM-score: 14.1)Zn_ribbon_5; zinc-ribbon domain (PF13719; HMM-score: 14.1)DUF7129; Domain of unknown function (DUF7129) (PF23455; HMM-score: 13.3)Zn_ribbon_Elf1; Transcription elongation factor Elf1 like (PF05129; HMM-score: 13.1)Zn_ribbon_TFIIS; Transcription factor S-II (TFIIS) (PF01096; HMM-score: 12.6)Ogr_Delta; Ogr/Delta-like zinc finger (PF04606; HMM-score: 11.9)no clan defined DUF7351; Domain of unknown function (DUF7351) (PF24042; HMM-score: 11.7)C2H2-zf (CL0361) zf-C2H2; Zinc finger, C2H2 type (PF00096; HMM-score: 11.4)Zn_Beta_Ribbon (CL0167) Zn_ribbon_PaaD; PaaD zinc beta ribbon domain (PF23451; HMM-score: 11.3)Zn_ribbon_4; zinc-ribbon domain (PF13717; HMM-score: 10.7)Zn_Ribbon_1; zinc-ribbon domain (PF13240; HMM-score: 9.7)no clan defined RUBY_RBDX; Rubrerythrin, rubredoxin-like domain (PF21349; HMM-score: 9.3)HEWD; HEWD domain (PF20576; HMM-score: 8.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: Zn2+
- effector: Deoxyribonucleotides
- genes regulated by NrdR, TF important in Deoxyribonucleotide biosynthesisRegPrecisetranscription units transferred from N315 data RegPrecise
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9776
- Cytoplasmic Membrane Score: 0.0025
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0198
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.016516
- TAT(Tat/SPI): 0.00067
- LIPO(Sec/SPII): 0.005624
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKCPKCNSTQSKVVDSRHADELNAIRRRRECENCGTRFTTFEHIEVSQLIVVKKDGTREQFSREKILNGLVRSCEKRPVRYQQLEDITNKVEWQLRDEGHTEVSSRDIGEHVMNLLMHVDQVSYVRFASVYKEFKDVDQLLASMQGILSENKRSDA
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_01791 < SAOUHSC_01792 < nrdRpredicted SigA promoter [3] : S707 < thrS < S708 < SAOUHSC_01789 < S709 < SAOUHSC_01791 < SAOUHSC_01792 < nrdR
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
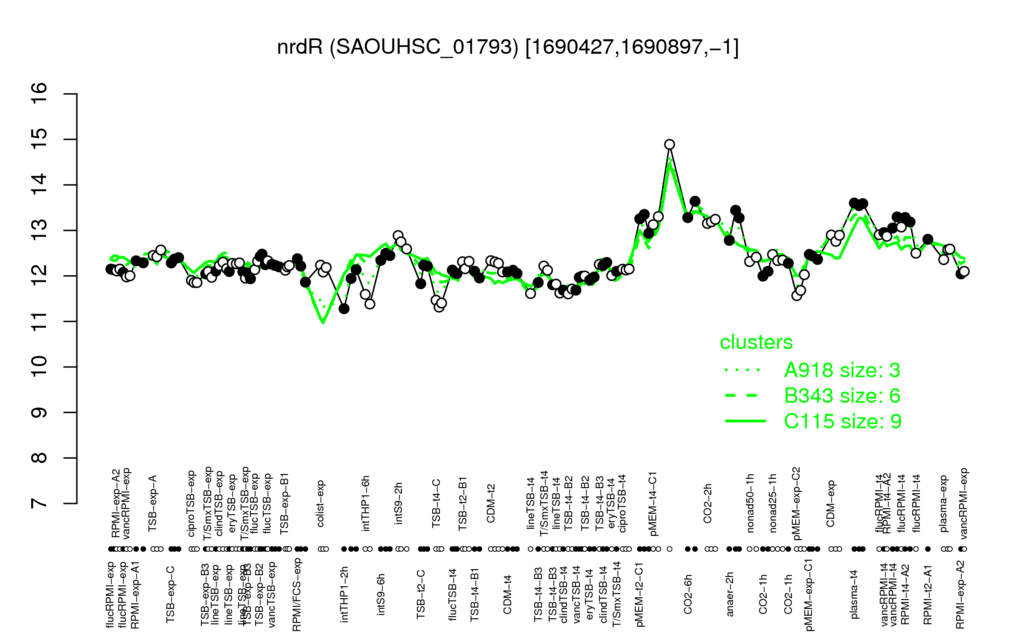 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Ilya Borovok, Batia Gorovitz, Michaela Yanku, Rachel Schreiber, Bertolf Gust, Keith Chater, Yair Aharonowitz, Gerald Cohen
Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation.
Mol Microbiol: 2004, 54(4);1022-35
[PubMed:15522084] [WorldCat.org] [DOI] (P p)
