⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01791
- pan locus tag?: SAUPAN004306000
- symbol: SAOUHSC_01791
- pan gene symbol?: dnaI
- synonym:
- product: primosomal protein DnaI
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3920432 NCBI
- RefSeq: YP_500295 NCBI
- BioCyc: G1I0R-1664 BioCyc
- MicrobesOnline: 1290209 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901ATGAAGCAATTTAAAAGTATAATTAACACGTCGCAGGACTTTGAAAAAAGAATAGAAAAG
ATAAAAAAAGAAGTAATCAATGACCCAGATGTTAAGCAATTTTTGGAAGCGCATCGAGCT
GAATTAACGAATGCTATGATTGATGAAGACTTAAATGTGTTACAAGAGTATAAAGATCAA
CAAAAACATTATGACGGTCATAAATTTGCTGATTGTCCAAATTTCGTAAAGGGGCATGTG
CCTGAGTTATATGTTGATAATAACCGAATTAAAATACGCTATTTACAATGCCCATGTAAA
ATCAAGTACGACGAAGAACGCTTTGAAGCTGAGCTAATTACATCTCATCATATGCAACGA
GATACTTTAAATGCCAAATTGAAAGATATTTATATGAATCATCGAGACCGTCTTGATGTA
GCTATGGCAGCAGATGATATTTGTACAGCAATAACTAATGGGGAACAAGTGAAAGGCCTT
TACCTTTATGGTCCATTTGGGACAGGTAAATCTTTTATTCTAGGTGCAATTGCGAATCAG
CTCAAATCTAAGAAGGTACGTTCGACAATTATTTATTTACCGGAATTTATTAGAACATTA
AAAGGTGGCTTTAAAGATGGTTCTTTTGAAAAGAAATTACATCGCGTAAGAGAAGCAAAC
ATTTTAATGCTTGATGATATTGGGGCTGAAGAAGTGACTCCATGGGTGAGAGATGAGGTA
ATTGGACCTTTGCTACATTATCGAATGGTTCATGAATTACCAACATTCTTTAGTTCTAAT
TTTGACTATAGTGAATTGGAACATCATTTAGCGATGACTCGTGATGGTGAAGAGAAGACT
AAAGCAGCACGTATTATTGAACGTGTCAAATCTTTGTCAACACCATACTTTTTATCAGGA
GAAAATTTCAGAAACAATTGA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
921
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01791
- symbol: SAOUHSC_01791
- description: primosomal protein DnaI
- length: 306
- theoretical pI: 7.15118
- theoretical MW: 35635.5
- GRAVY: -0.589543
⊟Function[edit | edit source]
- TIGRFAM: DNA metabolism DNA replication, recombination, and repair chromosomal replication initiator protein DnaA (TIGR00362; HMM-score: 38.3)and 6 moreDNA metabolism DNA replication, recombination, and repair DnaA regulatory inactivator Hda (TIGR03420; HMM-score: 21)AAA family ATPase, CDC48 subfamily (TIGR01243; HMM-score: 17.9)Protein fate Degradation of proteins, peptides, and glycopeptides proteasome ATPase (TIGR03689; EC 3.6.4.8; HMM-score: 17.3)26S proteasome subunit P45 family (TIGR01242; HMM-score: 15.5)DNA metabolism DNA replication, recombination, and repair orc1/cdc6 family replication initiation protein (TIGR02928; HMM-score: 13.7)DNA metabolism DNA replication, recombination, and repair Holliday junction DNA helicase RuvB (TIGR00635; EC 3.6.4.12; HMM-score: 11.5)
- TheSEED :
- Replicative DNA helicase loader DnaC (=BsDnaI)
- PFAM: no clan defined DnaI_N; Primosomal protein DnaI N-terminus (PF07319; HMM-score: 91.5)and 20 moreP-loop_NTPase (CL0023) Bac_DnaA; Bacterial DnaA ATPAse domain (PF00308; HMM-score: 32.1)IstB_IS21; IstB-like ATP binding protein (PF01695; HMM-score: 28.9)AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 24.2)AFG1_ATPase; AFG1-like ATPase (PF03969; HMM-score: 21)AAA_16; AAA ATPase domain (PF13191; HMM-score: 19.5)DUF5906; Family of unknown function (DUF5906) (PF19263; HMM-score: 18.1)PhoH; PhoH-like protein (PF02562; HMM-score: 16.1)AAA_22; AAA domain (PF13401; HMM-score: 15.5)PIF1; PIF1-like helicase (PF05970; HMM-score: 15.3)NPHP3_N; Nephrocystin 3, N-terminal (PF24883; HMM-score: 14.9)KAP_NTPase; KAP family P-loop domain (PF07693; HMM-score: 14)RNA_helicase; RNA helicase (PF00910; HMM-score: 13.5)ATPase_2; ATPase domain predominantly from Archaea (PF01637; HMM-score: 13.5)TsaE; Threonylcarbamoyl adenosine biosynthesis protein TsaE (PF02367; HMM-score: 13.2)ResIII; Type III restriction enzyme, res subunit (PF04851; HMM-score: 13.1)AAA_5; AAA domain (dynein-related subfamily) (PF07728; HMM-score: 12.8)AAA_7; P-loop containing dynein motor region (PF12775; HMM-score: 12.2)DUF815; Protein of unknown function (DUF815) (PF05673; HMM-score: 12.1)Cytidylate_kin; Cytidylate kinase (PF02224; HMM-score: 11.9)T3SS-hook (CL0424) Flg_hook; Flagellar hook-length control protein FliK (PF02120; HMM-score: 11.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9595
- Cytoplasmic Membrane Score: 0.0305
- Cell wall & surface Score: 0.0011
- Extracellular Score: 0.0089
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003359
- TAT(Tat/SPI): 0.000484
- LIPO(Sec/SPII): 0.000445
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKQFKSIINTSQDFEKRIEKIKKEVINDPDVKQFLEAHRAELTNAMIDEDLNVLQEYKDQQKHYDGHKFADCPNFVKGHVPELYVDNNRIKIRYLQCPCKIKYDEERFEAELITSHHMQRDTLNAKLKDIYMNHRDRLDVAMAADDICTAITNGEQVKGLYLYGPFGTGKSFILGAIANQLKSKKVRSTIIYLPEFIRTLKGGFKDGSFEKKLHRVREANILMLDDIGAEEVTPWVRDEVIGPLLHYRMVHELPTFFSSNFDYSELEHHLAMTRDGEEKTKAARIIERVKSLSTPYFLSGENFRNN
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_01791 < SAOUHSC_01792 < nrdRpredicted SigA promoter [4] : S707 < thrS < S708 < SAOUHSC_01789 < S709 < SAOUHSC_01791 < SAOUHSC_01792 < nrdR
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
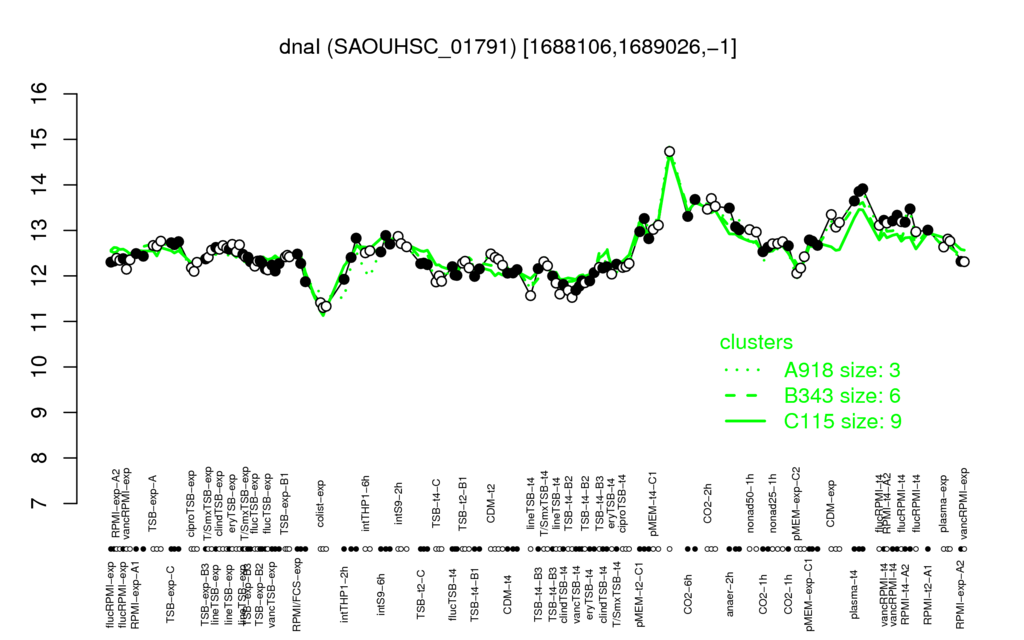 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Ilya Borovok, Batia Gorovitz, Michaela Yanku, Rachel Schreiber, Bertolf Gust, Keith Chater, Yair Aharonowitz, Gerald Cohen
Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation.
Mol Microbiol: 2004, 54(4);1022-35
[PubMed:15522084] [WorldCat.org] [DOI] (P p)Yan Li, Kenji Kurokawa, Luzia Reutimann, Hikaru Mizumura, Miki Matsuo, Kazuhisa Sekimizu
DnaB and DnaI temperature-sensitive mutants of Staphylococcus aureus: evidence for involvement of DnaB and DnaI in synchrony regulation of chromosome replication.
Microbiology (Reading): 2007, 153(Pt 10);3370-3379
[PubMed:17906136] [WorldCat.org] [DOI] (P p)
