Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00803
- pan locus tag?: SAUPAN002714000
- symbol: SAOUHSC_00803
- pan gene symbol?: rnr
- synonym:
- product: ribonuclease R
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3919366 NCBI
- RefSeq: YP_499359 NCBI
- BioCyc: G1I0R-752 BioCyc
- MicrobesOnline: 1289270 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341ATGAATTTAAAGCAATCTATAGAAGAGATTATTAATCAACCTGAATATGAACCTATGTCA
GTGTCAGATTTTCAAGATGCATTAGGTTTAAGCAGTGCCGACTCGTTTAGAGATTTAATT
AAGGTGCTTGTGGAGTTAGAACAATCAGGATTAATCGAACGTACAAAAACAGACAGATAC
CAAAAAAAGCATAGTTATAGAGGTCAATCAAAATTGATAAAAGGAACGTTAAGTCAAAAT
AAAAAAGGCTTTGCATTCTTAAGACCTGAAGATGAGGATATGGAAGATATATTTATTCCC
CCGACGAAAATTAATCGTGCCTTGGATGGAGATACTGTTATTGTAGAAATCCATCAATCA
AAAGGTGAACATAAAGGTAAAATCGAAGGGGAAGTTAAGTCGATTGAGAAGCATTCTGTA
ACTCAAGTTGTTGGTACGTATAGTGAAGCTAGACATTTTGGCTTTGTTATTCCGGATGAT
AAACGTATTATGCAAGATATTTTCATTCCTAAAGGTCAAAGTTTAGGCGCAGTCGATGGT
CATAAGGTACTTGTACAAATTACTAAGTATGCTGATGGTTCAGATAATCCAGAAGGACAT
ATTTCTGCTATTTTAGGACATAAAAATGATCCTGGCGTAGATATTTTATCTATTATCTAT
CAACATGGCATAGAAATTGAATTTCCTGATGAAGTGTTACAAGAAGCTGAAGCAGTACCT
GATCATATTGAAAATACTGAAATTAAAGGCCGTCATGATTTACGTGATGAATTGACAATC
ACAATTGATGGTGCTGATGCTAAAGACTTAGATGACGCAATTAGTGTTAAAAAGTTAGCG
AACGGTAATACGCAATTAACTGTAAGTATTGCTGATGTCAGCTATTATGTAACAGAAGGT
TCTGCATTGGATAAAGAGGCATATGATAGAGCGACAAGTGTATATCTTGTTGACCGTGTA
ATTCCAATGATTCCACATCGATTAAGTAATGGTATTTGTTCATTGAATCCTAATGTTGAT
CGTTTAACTCTAAGCTGTCGCATGGAAATCGATGCTAGTGGTCGCGTTGTTAAACATGAA
ATTTTTGATAGTGTTATACATTCTGATTATCGAATGACGTATGATGCGGTAAATCAGATT
ATTACTGAAAAGGATCCTAACATTCGCGAACAATATAATGAAATTACGCCTATGCTAGAT
TTAGCACAAGATTTATCTAATCGTTTGATTCAAATGAGAAAACGACGTGGTGAAATCGAT
TTTGATATTAGTGAAGCAAAAGTATTAGTTAACGAAGACGGTATACCAACAGATGTTCAA
TTAAGACAACGTGGCGAGGGTGAACGTCTAATTGAATCATTTATGTTAATTGCAAATGAA
ACAGTTGCTGAACATTTTAGTAAGTTAGATGTACCTTTTATTTACCGAGTGCATGAGCAA
CCTAAATCAGATCGCTTAAGACAATTCTTTGATTTTATTACAAACTTTGGCATCATGATT
AAGGGTACTGGCGAAGATATTCATCCAACAACACTTCAAAAGGTTCAAGAAGAAGTAGAA
GGTCGACCTGAACAAATGGTCATTTCAACAATGATGTTGCGTTCAATGCAACAAGCGCAT
TATGATGATGTGAACTTGGGACATTTTGGCTTATCAGCTGAATATTATACGCATTTTACA
TCACCAATTAGACGTTATCCTGATTTAACAGTTCATCGTTTAATCCGTAAGTATTTAATT
GAGAAATCAATGGATAACAAAGAAGTGAAGCGTTGGGAAGACAAATTGCCTGAGTTAGCT
GAACATACTTCTAAACGTGAACGTCGTGCTATTGAGGCAGAACGTGATACTGATGAATTG
AAAAAAGCAGAATATATGATTCAACATATTGGTGATGAATTTGAAGGTATTGTCAGCTCA
GTAGCTAACTTCGGTATGTTCATTGAATTGCCAAATACGATAGAAGGTATGGTTCATATT
GCGAATATGACTGATGATTATTACCGTTTTGAAGAGCGTCAAATGGCATTAATTGGTGAG
CGTCAAGCTAAAGTATTTAGAATTGGTGACACAGTTAAGGTTAAAGTGACGCATGTTGAT
GTAGATGAACGATTAATTGATTTTCAAATTGTAGGTATGCCTTTACCGAAAAATGATCGA
TCACAGCGCCCAGCGCGAGGTAAGACAATTCAAGCCAAAACGCGTGGTAAATCATTAGAT
AAATCAAAATCTGATGATAAGGGTCGTAAGAAAAAAGGTAAGCAACGTAAAGGTAAAAAC
CAACGTAATAATGATAAATCAGGTAATAGTAAGCATAAGCCATTTTATAAAGATAAAAGT
GTGAAAAAGAAAGCACGTCGTAAGAAAAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2373
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00803
- symbol: SAOUHSC_00803
- description: ribonuclease R
- length: 790
- theoretical pI: 6.65179
- theoretical MW: 90434.9
- GRAVY: -0.672278
⊟Function[edit | edit source]
- reaction: EC 3.1.13.1? ExPASyExoribonuclease II Exonucleolytic cleavage in the 3'- to 5'-direction to yield nucleoside 5'-phosphates
- TIGRFAM: Transcription Degradation of RNA ribonuclease R (TIGR02063; EC 3.1.-.-; HMM-score: 961.8)and 6 moreTranscription Degradation of RNA VacB and RNase II family 3'-5' exoribonucleases (TIGR00358; EC 3.1.13.1; HMM-score: 733.8)Transcription Degradation of RNA exoribonuclease II (TIGR02062; EC 3.1.13.1; HMM-score: 206.2)Transcription DNA-dependent RNA polymerase DNA-directed RNA polymerase (TIGR00448; EC 2.7.7.6; HMM-score: 40.7)Protein synthesis Ribosomal proteins: synthesis and modification ribosomal protein bS1 (TIGR00717; HMM-score: 30.4)Transcription Degradation of RNA polyribonucleotide nucleotidyltransferase (TIGR03591; EC 2.7.7.8; HMM-score: 25.4)guanosine pentaphosphate synthetase I/polyribonucleotide nucleotidyltransferase (TIGR02696; EC 2.7.-.-,2.7.7.8; HMM-score: 14.7)
- TheSEED :
- 3'-to-5' exoribonuclease RNase R
- PFAM: no clan defined RNB; RNB domain (PF00773; HMM-score: 382.8)and 13 moreOB (CL0021) CSD2; Cold shock domain (PF17876; HMM-score: 128.2)OB_RNB; Ribonuclease B OB domain (PF08206; HMM-score: 102)S1; S1 RNA binding domain (PF00575; HMM-score: 61.2)S1CSD-TOTE-2; S1/CSD-like domain 2 of the TOTE conflict systems (PF22707; HMM-score: 25.7)OB_HELZ2; 3'-5' exoribonuclease HELZ2, OB-fold domain (PF25049; HMM-score: 25.7)OB_Dis3; Dis3-like cold-shock domain 2 (CSD2) (PF17849; HMM-score: 19.9)OB_RNR_1st; Ribonuclease R, OB first domain, N-terminal (PF22896; HMM-score: 17.4)Rho_RNA_bind; Rho termination factor, RNA-binding domain (PF07497; HMM-score: 17.2)CSD; 'Cold-shock' DNA-binding domain (PF00313; HMM-score: 16.3)RNase_II_C_S1; RNase II-type exonuclease C-terminal S1 domain (PF18614; HMM-score: 15.2)PDDEXK (CL0236) TBPIP_N; TBP-interacting protein N-terminus (PF15517; HMM-score: 14.8)OB (CL0021) S1_RRP5; RRP5 S1 domain (PF23459; HMM-score: 14.5)HTH (CL0123) MarR; MarR family (PF01047; HMM-score: 13)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9921
- Cytoplasmic Membrane Score: 0.006
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0019
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003415
- TAT(Tat/SPI): 0.001061
- LIPO(Sec/SPII): 0.000359
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNLKQSIEEIINQPEYEPMSVSDFQDALGLSSADSFRDLIKVLVELEQSGLIERTKTDRYQKKHSYRGQSKLIKGTLSQNKKGFAFLRPEDEDMEDIFIPPTKINRALDGDTVIVEIHQSKGEHKGKIEGEVKSIEKHSVTQVVGTYSEARHFGFVIPDDKRIMQDIFIPKGQSLGAVDGHKVLVQITKYADGSDNPEGHISAILGHKNDPGVDILSIIYQHGIEIEFPDEVLQEAEAVPDHIENTEIKGRHDLRDELTITIDGADAKDLDDAISVKKLANGNTQLTVSIADVSYYVTEGSALDKEAYDRATSVYLVDRVIPMIPHRLSNGICSLNPNVDRLTLSCRMEIDASGRVVKHEIFDSVIHSDYRMTYDAVNQIITEKDPNIREQYNEITPMLDLAQDLSNRLIQMRKRRGEIDFDISEAKVLVNEDGIPTDVQLRQRGEGERLIESFMLIANETVAEHFSKLDVPFIYRVHEQPKSDRLRQFFDFITNFGIMIKGTGEDIHPTTLQKVQEEVEGRPEQMVISTMMLRSMQQAHYDDVNLGHFGLSAEYYTHFTSPIRRYPDLTVHRLIRKYLIEKSMDNKEVKRWEDKLPELAEHTSKRERRAIEAERDTDELKKAEYMIQHIGDEFEGIVSSVANFGMFIELPNTIEGMVHIANMTDDYYRFEERQMALIGERQAKVFRIGDTVKVKVTHVDVDERLIDFQIVGMPLPKNDRSQRPARGKTIQAKTRGKSLDKSKSDDKGRKKKGKQRKGKNQRNNDKSGNSKHKPFYKDKSVKKKARRKKK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_01786 (infC) translation initiation factor IF-3 [4] (data from MRSA252) SAOUHSC_02509 (rplB) 50S ribosomal protein L2 [4] (data from MRSA252) SAOUHSC_02512 (rplC) 50S ribosomal protein L3 [4] (data from MRSA252) SAOUHSC_02511 (rplD) 50S ribosomal protein L4 [4] (data from MRSA252) SAOUHSC_02500 (rplE) 50S ribosomal protein L5 [4] (data from MRSA252) SAOUHSC_02496 (rplF) 50S ribosomal protein L6 [4] (data from MRSA252) SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [4] (data from MRSA252) SAOUHSC_00521 (rplL) 50S ribosomal protein L7/L12 [4] (data from MRSA252) SAOUHSC_02492 (rplO) 50S ribosomal protein L15 [4] (data from MRSA252) SAOUHSC_02505 (rplP) 50S ribosomal protein L16 [4] (data from MRSA252) SAOUHSC_01211 (rplS) 50S ribosomal protein L19 [4] (data from MRSA252) SAOUHSC_01757 (rplU) 50S ribosomal protein L21 [4] (data from MRSA252) SAOUHSC_02507 (rplV) 50S ribosomal protein L22 [4] (data from MRSA252) SAOUHSC_02510 (rplW) 50S ribosomal protein L23 [4] (data from MRSA252) SAOUHSC_01829 (rpsD) 30S ribosomal protein S4 [4] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [4] (data from MRSA252) SAOUHSC_00348 (rpsF) 30S ribosomal protein S6 [4] (data from MRSA252) SAOUHSC_02477 (rpsI) 30S ribosomal protein S9 [4] (data from MRSA252) SAOUHSC_02503 (rpsQ) 30S ribosomal protein S17 [4] (data from MRSA252) SAOUHSC_02508 (rpsS) 30S ribosomal protein S19 [4] (data from MRSA252) SAOUHSC_00284 5'-nucleotidase [4] (data from MRSA252) SAOUHSC_00528 30S ribosomal protein S7 [4] (data from MRSA252) SAOUHSC_01035 hypothetical protein [4] (data from MRSA252) SAOUHSC_01391 hypothetical protein [4] (data from MRSA252) SAOUHSC_01490 DNA-binding protein HU [4] (data from MRSA252) SAOUHSC_01814 hypothetical protein [4] (data from MRSA252) SAOUHSC_02133 nicotinate phosphoribosyltransferase [4] (data from MRSA252) SAOUHSC_02316 DEAD-box ATP dependent DNA helicase [4] (data from MRSA252) SAOUHSC_02486 30S ribosomal protein S11 [4] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_00802 > SAOUHSC_00803 > smpBpredicted SigA promoter [5] : S327 > SAOUHSC_00800 > secG > S328 > SAOUHSC_00802 > SAOUHSC_00803 > smpB
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [5]
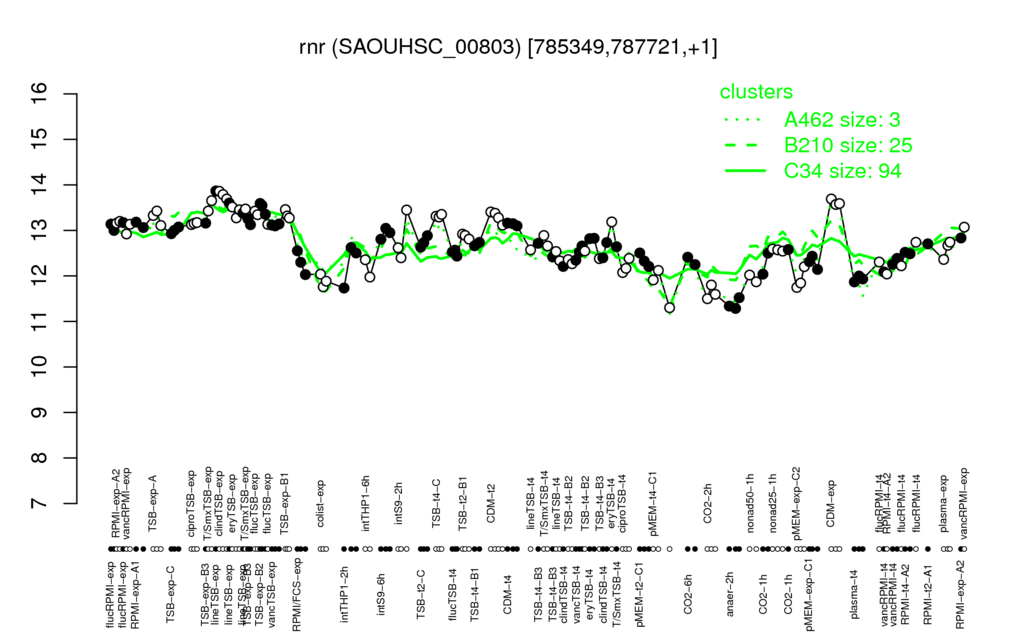 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 4.20 4.21 4.22 4.23 4.24 4.25 4.26 4.27 4.28 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 5.0 5.1 5.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
