⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00515
- pan locus tag?: SAUPAN002302000
- symbol: SAOUHSC_00515
- pan gene symbol?: sigH
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00515
- symbol: SAOUHSC_00515
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 516850..517419
- length: 570
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920369 NCBI
- RefSeq: YP_499088 NCBI
- BioCyc: G1I0R-486 BioCyc
- MicrobesOnline: 1288998 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541TTGAAATACGATTTGACAACTCAAGACAGTACAATCAAACGTAACAATGCAATAAATGAT
AAAGACTTCGAAAAGTTAGTAATGGATCTACAACCATTAATTATTCGACGCATCAAAACA
TTTGGATTTAATCATTATGATTTAGAAGACTTATATCAAGAAATACTTATACGGATGTAT
AGGTCGGTCCAAACATTTGATTTTAGTGGAGAGCAGCCTTTCACAAATTATGTTCAATGT
TTAATTACGTCTGTAAAGTATGATTATTTGAGAAAATATTTAGCTACAAATAAAAGAATG
GATAATTTGATTAATGAATATAGAGTTACGTATCCATGTGCAATAAAGCGTTTTGATGTT
GAAAACAATTATTTGAATAAATTAGCAATTAAAGAGTTGATTTGTCAGTTTAAGTATTTG
AGTGCATTTGAAAAAGATGTCATGTATTTAATGTGTGAACAATATAAGCCGAGAGAAATT
GCTCAATTGATGCATGTAAAAGAGAAAGTGATTTATAATGCCATACAACGATGTAAAAAT
AAAATAAAACGTTATTTCAAAATGATTTGA60
120
180
240
300
360
420
480
540
570
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00515
- symbol: SAOUHSC_00515
- description: hypothetical protein
- length: 189
- theoretical pI: 9.43118
- theoretical MW: 23028.8
- GRAVY: -0.478836
⊟Function[edit | edit source]
- TIGRFAM: RNA polymerase sigma factor, sigma-70 family (TIGR02937; HMM-score: 53.2)and 17 moreRNA polymerase sigma-70 factor, Bacteroides expansion family 1 (TIGR02985; HMM-score: 29)Cellular processes Sporulation and germination RNA polymerase sigma-H factor (TIGR02859; HMM-score: 27.8)Transcription Transcription factors RNA polymerase sigma-H factor (TIGR02859; HMM-score: 27.8)RNA polymerase sigma-70 factor, sigma-B/F/G subfamily (TIGR02980; HMM-score: 25.2)RNA polymerase sigma-70 factor, TIGR02952 family (TIGR02952; HMM-score: 25.1)RNA polymerase sigma-70 factor, Rhodopirellula/Verrucomicrobium family (TIGR02989; HMM-score: 25.1)Cellular processes Sporulation and germination RNA polymerase sigma-F factor (TIGR02885; HMM-score: 23.8)Transcription Transcription factors RNA polymerase sigma-F factor (TIGR02885; HMM-score: 23.8)RNA polymerase sigma factor RpoE (TIGR02939; HMM-score: 21)RNA polymerase sigma factor, FliA/WhiG family (TIGR02479; HMM-score: 19.9)RNA polymerase sigma factor, SigM family (TIGR02950; HMM-score: 19.8)RNA polymerase sigma-70 factor, Planctomycetaceae-specific subfamily 1 (TIGR02984; HMM-score: 19.4)RNA polymerase sigma-W factor (TIGR02948; HMM-score: 16.4)Cellular processes Sporulation and germination RNA polymerase sigma-G factor (TIGR02850; HMM-score: 15.4)Transcription Transcription factors RNA polymerase sigma-G factor (TIGR02850; HMM-score: 15.4)RNA polymerase sigma-70 factor, sigma-E family (TIGR02983; HMM-score: 15.4)RNA polymerase sigma-70 factor, TIGR02954 family (TIGR02954; HMM-score: 12.9)
- TheSEED :
- RNA polymerase sporulation specific sigma factor SigH
Dormancy and Sporulation Dormancy and Sporulation - no subcategory Sporulation-associated proteins with broader functions RNA polymerase sporulation specific sigma factor SigHand 2 more - PFAM: HTH (CL0123) Sigma70_r2; Sigma-70 region 2 (PF04542; HMM-score: 47)and 6 moreSigma70_r4_2; Sigma-70, region 4 (PF08281; HMM-score: 17.5)UPF0122; Putative helix-turn-helix protein, YlxM / p13 like (PF04297; HMM-score: 13.7)HTH_23; Homeodomain-like domain (PF13384; HMM-score: 13.6)Terminase_5; Putative ATPase subunit of terminase (gpP-like) (PF06056; HMM-score: 13.2)GerE; Bacterial regulatory proteins, luxR family (PF00196; HMM-score: 12.4)HTH_29; Winged helix-turn helix (PF13551; HMM-score: 12.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.8328
- Cytoplasmic Membrane Score: 0.0634
- Cell wall & surface Score: 0.0017
- Extracellular Score: 0.1022
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.001521
- TAT(Tat/SPI): 0.000208
- LIPO(Sec/SPII): 0.000353
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKYDLTTQDSTIKRNNAINDKDFEKLVMDLQPLIIRRIKTFGFNHYDLEDLYQEILIRMYRSVQTFDFSGEQPFTNYVQCLITSVKYDYLRKYLATNKRMDNLINEYRVTYPCAIKRFDVENNYLNKLAIKELICQFKYLSAFEKDVMYLMCEQYKPREIAQLMHVKEKVIYNAIQRCKNKIKRYFKMI
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [2] : S171 > S172 > SAOUHSC_00507 > SAOUHSC_00508 > S173 > S174 > gltX > S175 > S176 > SAOUHSC_00510 > cysS > SAOUHSC_00512 > SAOUHSC_00513 > SAOUHSC_00514 > S177 > SAOUHSC_00515 > S178 > S179 > secE > SAOUHSC_00517 > S180 > rplK > S181 > rplA > S182predicted SigA promoter [2] : S174 > gltX > S175 > S176 > SAOUHSC_00510 > cysS > SAOUHSC_00512 > SAOUHSC_00513 > SAOUHSC_00514 > S177 > SAOUHSC_00515 > S178 > S179 > secE > SAOUHSC_00517 > S180 > rplK > S181 > rplA > S182 > S183 > rplJ > rplLpredicted SigA promoter [2] : S175 > S176 > SAOUHSC_00510 > cysS > SAOUHSC_00512 > SAOUHSC_00513 > SAOUHSC_00514 > S177 > SAOUHSC_00515 > S178 > S179 > secE > SAOUHSC_00517 > S180 > rplK > S181 > rplA > S182
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [2]
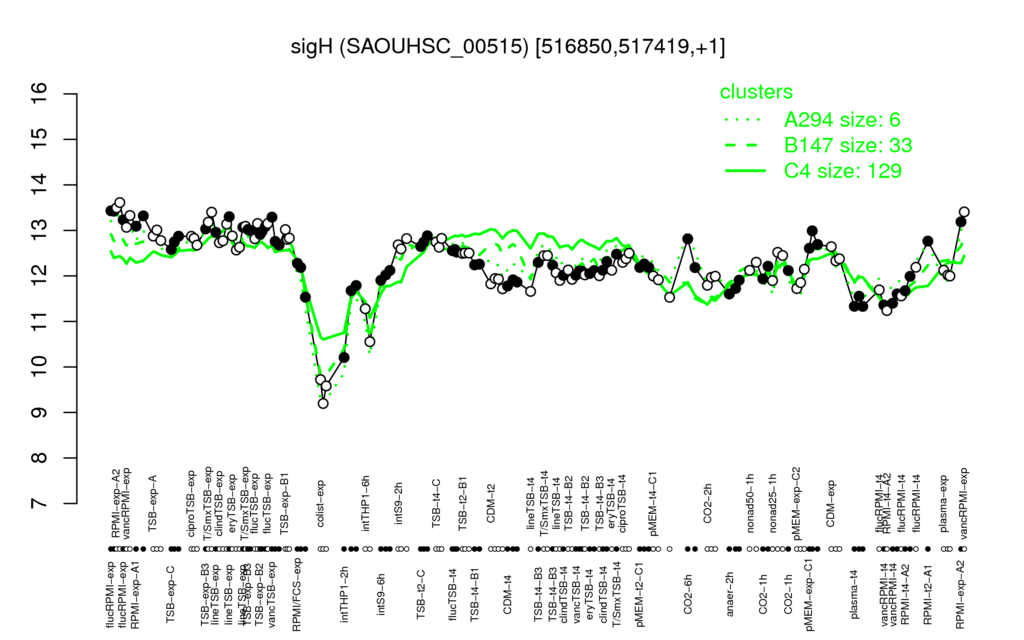 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Liang Tao, Xiaoqian Wu, Baolin Sun
Alternative sigma factor sigmaH modulates prophage integration and excision in Staphylococcus aureus.
PLoS Pathog: 2010, 6(5);e1000888
[PubMed:20485515] [WorldCat.org] [DOI] (I e) - ↑ 2.0 2.1 2.2 2.3 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Kazuya Morikawa, Yumiko Inose, Hideyuki Okamura, Atsushi Maruyama, Hideo Hayashi, Kunio Takeyasu, Toshiko Ohta
A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes.
Genes Cells: 2003, 8(8);699-712
[PubMed:12875655] [WorldCat.org] [DOI] (P p)Liang Tao, Xiaoqian Wu, Baolin Sun
Alternative sigma factor sigmaH modulates prophage integration and excision in Staphylococcus aureus.
PLoS Pathog: 2010, 6(5);e1000888
[PubMed:20485515] [WorldCat.org] [DOI] (I e)Kazuya Morikawa, Aya J Takemura, Yumiko Inose, Melody Tsai, Le Thuy Nguyen Thi, Toshiko Ohta, Tarek Msadek
Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus.
PLoS Pathog: 2012, 8(11);e1003003
[PubMed:23133387] [WorldCat.org] [DOI] (I p)
