⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02163
- pan locus tag?: SAUPAN005011000
- symbol: SAOUHSC_02163
- pan gene symbol?: hlb-1
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02163
- symbol: SAOUHSC_02163
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 2031741..2031938
- length: 198
- essential: no DEG other strains
- comment: SAOUHSC_02163 is the N-terminal part of the hlb gene which is split by insertion of the Φ13 prophage; complete protein: UniProt Q2FWP1
⊟Accession numbers[edit | edit source]
- Gene ID: 3921858 NCBI
- RefSeq: YP_500651 NCBI
- BioCyc: G1I0R-3581 BioCyc
- MicrobesOnline: 1290607 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181ATGGTGAAAAAAACAAAATCCAATTCACTAAAAAAAGTTGCAACACTTGCATTAGCAAAT
TTATTATTAGTTGGTGCACTTACTGACAATAGTGCCAAAGCCGAATCTAAGAAAGATGAT
ACTGATTTGAAGTTAGTTAGTCATAACGTTTATATGTTATCGACCGTTTTGTATCCAAAC
TGGAGACTTTTAACATAA60
120
180
198
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02163
- symbol: SAOUHSC_02163
- description: hypothetical protein
- length: 65
- theoretical pI: 10.4211
- theoretical MW: 7175.41
- GRAVY: -0.0476923
⊟Function[edit | edit source]
- reaction: EC 3.1.4.3? ExPASyPhospholipase C A phosphatidylcholine + H2O = 1,2-diacyl-sn-glycerol + phosphocholine
- TIGRFAM: Cellular processes Pathogenesis sphingomyelin phosphodiesterase (TIGR03395; EC 3.1.4.12; HMM-score: 14)
- TheSEED :
- Beta-hemolysin
- PFAM:
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 8.8
- Cellwall Score: 0.22
- Extracellular Score: 0.98
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.0007
- Cell wall & surface Score: 0.0017
- Extracellular Score: 0.9976
- LocateP: Secretory(released) (with CS)
- Prediction by SwissProt Classification: Extracellular
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: -0.17
- Signal peptide possibility: 1
- N-terminally Anchored Score: -2
- Predicted Cleavage Site: NSAKAESK
- SignalP: Signal peptide SP(Sec/SPI) length 34 aa
- SP(Sec/SPI): 0.959976
- TAT(Tat/SPI): 0.010867
- LIPO(Sec/SPII): 0.020042
- Cleavage Site: CS pos: 34-35. AKA-ES. Pr: 0.9130
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MVKKTKSNSLKKVATLALANLLLVGALTDNSAKAESKKDDTDLKLVSHNVYMLSTVLYPNWRLLT
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [1] : SAOUHSC_02163 > S848 > S849 > S850 > SAOUHSC_02164 > S851 > SAOUHSC_02166
⊟Regulation[edit | edit source]
- regulator: CodY* (repression) regulon
CodY* (TF) important in Amino acid metabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
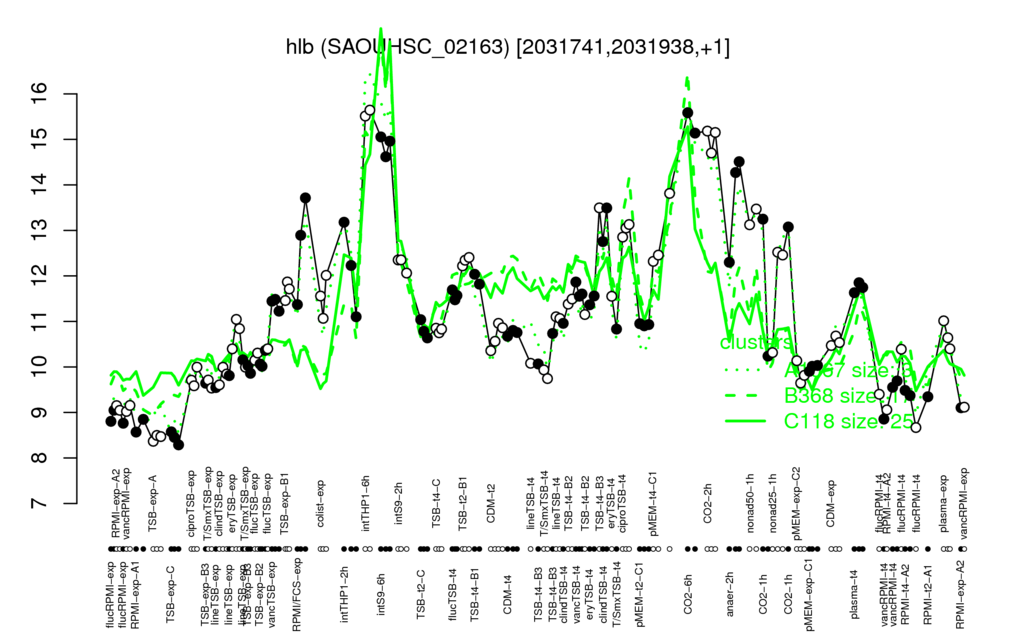 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Mark J J B Sibbald, Theresa Winter, Magdalena M van der Kooi-Pol, G Buist, E Tsompanidou, Tjibbe Bosma, Tina Schäfer, Knut Ohlsen, Michael Hecker, Haike Antelmann, Susanne Engelmann, Jan Maarten van Dijl
Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus.
J Bacteriol: 2010, 192(14);3788-800
[PubMed:20472795] [WorldCat.org] [DOI] (I p)K Dziewanowska, V M Edwards, J R Deringer, G A Bohach, D J Guerra
Comparison of the beta-toxins from Staphylococcus aureus and Staphylococcus intermedius.
Arch Biochem Biophys: 1996, 335(1);102-8
[PubMed:8914839] [WorldCat.org] [DOI] (P p)
