Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01857
- pan locus tag?: SAUPAN004408000
- symbol: SAOUHSC_01857
- pan gene symbol?: ftsK
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01857
- symbol: SAOUHSC_01857
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1763159..1766983
- length: 3825
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920535 NCBI
- RefSeq: YP_500361 NCBI
- BioCyc: G1I0R-1727 BioCyc
- MicrobesOnline: 1290275 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601
3661
3721
3781ATGAGCTGGTTTGATAAATTATTCGGCGAAGATAATGATTCAAATGATGACTTGATTCAT
AGAAAGAAAAAAAGACGTCAAGAATCACAAAATATAGATAACGATCATGACTCATTACTG
CCTCAAAATAATGATATTTATAGTCGTCCGAGGGGAAAATTCCGTTTTCCTATGAGCGTA
GCTTATGAAAATGAAAATGTTGAACAATCTGCAGATACTATTTCAGATGAAAAAGAACAA
TACCATCGAGACTATCGCAAACAAAGCCACGATTCTCGTTCACAAAAACGACATCGCCGT
AGAAGAAATCAAACAACTGAAGAACAAAATTATAGTGAACAACGTGGGAATTCTAAAATA
TCACAGCAAAGTATAAAATATAAAGATCATTCACATTACCATACGAATAAGCCAGGTACA
TATGTTTCTGCAATTAATGGTATTGAGAAGGAAACGCACAAGCCAAAAACACATAATATG
TATTCTAATAATACAAATCATCGTGCTAAAGATTCAACTCCAGATTATCACAAAGAAAGT
TTCAAGACTTCAGAGGTACCGTCAGCTATTTTTGGCACAATGAAACCTAAAAAGTTAGAA
AATGGTCGTATCCCTGTAAGTAAACCTTCAGAAAAAGTTGAGTCAGATAAACAAAAATAT
GATAAATATGTAGCTAAGACGCAAACGTCTCAAAATAAACAATTAGAACAAGAAAAACAA
AATGATAGTGTTGTCAAACAAGGAACTGCATCTAAATCATCTGATGAAAATGTATCATCA
ACAACAAAATCAATGCCTAATTATTCAAAAGTTGATAATACTATCAAAATTGAAAATATT
TATGCTTCACAAATTGTTGAAGAAATTAGACGTGAACGAGAACGTAAAGTGCTTCAAAAG
CGTCGATTTAAAAAAGCGTTGCAACAAAAGCGTGAAGAACATAAAAACGAAGAGCAAGAT
GCAATACAACGTGCAATTGATGAAATGTATGCTAAACAAGCGGAACGCTATGTTGGTGAT
AGTTCATTAAATGATGATAGTGACTTAACAGATAATAGTACAGATGCTAGTCAGCTTCAT
ACAAATGGCATAGAGAATGAAACTGTATCAAATGATGAAAATAAACAAGCGTCAATACAA
AATGAAGACACTAATGACACTCATGTAGATGAAAGTCCATACAATTATGAGGAAGTTAGT
TTGAATCAAGTATCGACAACAAAACAATTGTCAGATGATGAAGTTACGGTTTCGAATGTA
ACGTCTCAACATCAATCAGCACTACAACATAACGTTGAAGTAAATGATAAAGATGAACTA
AAAAATCAATCCAGATTAATTGCTGATTCAGAAGAAGATGGAGCAACGAATAAAGAAGAA
TATTCAGGAAGTCAAATCGATGATGCAGAATTTTATGAATTAAATGATACAGAAGTAGAT
GAGGATACTACTTCAAATATCGAAGATAATACCAATAGAAACGCGTCTGAAATGCATGTA
GACGCTCCTAAAACGCAAGAGTACGCAGTAACTGAATCTCAAGTAAATAATATCGATAAA
ACGGTTGATAATGAAATTGAATTAGCACCGCGTCATAAAAAAGATGACCAAACAAACTTA
AGTGTCAACTCATTGAAAACGAATGATGTGAATGATAATCATGTTGTGGAAGATTCAAGC
ATGAATGAAATAGAAAAGAATAACGCAGAAATTACAGAAAATGTGCAAAACGAAGCAGCT
GAAAGTGAACAAAATGTCGAAGAGAAAACTATTGAAAACGTAAATCCAAAGAAACAGACT
GAAAAGGTTTCAACTTTAAGTAAAAGACCATTTAATGTTGTCATGACGCCATCTGATAAA
AAGCGTATGATGGATCGTAAAAAGCATTCAAAAGTCAATGTGCCTGAATTAAAGCCTGTA
CAAAGTAAGCAAGCTGTGAGTGAAAGAATGCCTGCGAGTCAAGCCACACCATCATCAAGA
TCTGATTCACAAGAGTCAAATACAAATGCATATAAAACAAATAATATGACATCAAACAAT
GTTGAGAACAATCAACTTATTGGTCATGCAGAAACAGAAAATGATTATCAAAATGCACAA
CAATATTCAGAGCAGAAACCTTCTGTTGATTCAACTCAAACGGAAATATTTGAAGAAAGT
CAAGATGATAATCAATTGGAAAATGAGCAAGTTGATCAATCAACTTCGTCTTCAGTTTCA
GAAGTAAGCGACATAACTGAAGAAAGCGAAGAAACAACACATCCAAACAATACTAGTGGA
CAACAAGATAATGATGATCAACAAAAAGATTTACAGTCATCATTTTCAAATAAAAATGAA
GATACAGCTAATGAAAATAGACCTCGGACGAACCAACAAGATGTTGCAACAAATCAAGCT
GTACAAACATCTAAGCCGATGATTCGTAAAGGCCCAAATATTAAATTGCCAAGTGTTTCA
TTACTAGAAGAACCACAAGTTATTGAGTCGGACGAGGACTGGATTACAGATAAAAAGAAA
GAACTGAATGACGCATTATTTTACTTTAATGTACCTGCAGAAGTACAAGATGTAACTGAA
GGTCCAAGTGTTACAAGATTTGAATTATCAGTTGAAAAAGGTGTTAAAGTTTCAAGAATT
ACGGCATTACAAGATGACATTAAAATGGCATTGGCAGCGAAAGATATTCGTATAGAAGCG
CCTATTCCAGGAACTAGTCGTGTTGGTATTGAAGTTCCGAACCAAAATCCAACGACAGTC
AACTTACGTTCTATTATTGAATCTCCAAGTTTTAAAAATGCTGAATCTAAATTAACAGTT
GCGATGGGGTATAGAATTAATAATGAACCATTACTTATGGATATTGCTAAAACGCCACAC
GCACTAATTGCAGGTGCAACTGGATCAGGGAAATCAGTTTGTATCAATAGTATTTTGATG
TCTTTACTATATAAAAATCATCCTGAGGAATTAAGATTATTACTTATCGATCCAAAAATG
GTTGAATTAGCTCCTTATAATGGTTTGCCACATTTAGTTGCACCGGTAATTACAGATGTC
AAAGCAGCTACACAGAGTTTAAAATGGGCCGTAGAAGAAATGGAACGACGTTATAAGTTA
TTTGCACATTACCATGTACGTAATATAACAGCATTTAACAAAAAAGCACCATATGATGAA
AGAATGCCAAAAATTGTCATTGTAATTGATGAGTTGGCTGATTTAATGATGATGGCTCCG
CAAGAAGTTGAACAGTCTATTGCTAGAATTGCTCAAAAAGCGAGAGCATGTGGTATTCAT
ATGTTAGTAGCTACGCAAAGACCATCTGTCAATGTAATTACAGGTTTAATTAAAGCCAAC
ATACCAACAAGAATTGCATTTATGGTATCATCAAGTGTAGATTCGAGAACGATATTAGAC
AGTGGTGGAGCAGAACGCTTGTTAGGATATGGCGATATGTTATATCTTGGTAGCGGTATG
AATAAACCGATTAGAGTTCAAGGTACATTTGTTTCTGATGACGAAATTGATGATGTTGTT
GATTTTATCAAACAACAAAGAGAACCGGACTATCTATTTGAAGAAAAAGAATTGTTGAAA
AAAACACAAACACAATCACAAGATGAATTATTTGATGATGTTTGTGCATTTATGGTTAAT
GAAGGACATATTTCAACATCATTAATCCAAAGACATTTCCAAATTGGCTATAATAGAGCA
GCAAGAATTATCGATCAATTAGAGCAACTCGGTTATGTTTCGAGTGCTAATGGTTCAAAA
CCAAGGGATGTTTATGTTACGGAAGCAGATTTAAATAAAGAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3660
3720
3780
3825
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01857
- symbol: SAOUHSC_01857
- description: hypothetical protein
- length: 1274
- theoretical pI: 4.84013
- theoretical MW: 144655
- GRAVY: -1.01593
⊟Function[edit | edit source]
- TIGRFAM: Protein fate Protein and peptide secretion and trafficking type VII secretion protein EssC (TIGR03928; HMM-score: 129.6)Protein fate Protein and peptide secretion and trafficking type VII secretion protein EccCa (TIGR03924; HMM-score: 107.4)and 1 moreProtein fate Protein and peptide secretion and trafficking type VII secretion protein EccCb (TIGR03925; HMM-score: 87.1)
- TheSEED :
- Cell division protein FtsK
- PFAM: P-loop_NTPase (CL0023) FtsK_SpoIIIE; FtsK/SpoIIIE family (PF01580; HMM-score: 221.4)and 5 moreno clan defined FtsK_alpha; FtsK alpha domain (PF17854; HMM-score: 102.4)HTH (CL0123) FtsK_gamma; Ftsk gamma domain (PF09397; HMM-score: 95)P-loop_NTPase (CL0023) DEAD; DEAD/DEAH box helicase (PF00270; HMM-score: 16.1)AAA_16; AAA ATPase domain (PF13191; HMM-score: 14.8)AAA_22; AAA domain (PF13401; HMM-score: 14.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 1.05
- Cytoplasmic Membrane Score: 8.78
- Cellwall Score: 0.08
- Extracellular Score: 0.09
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.7901
- Cytoplasmic Membrane Score: 0.0866
- Cell wall & surface Score: 0.0276
- Extracellular Score: 0.0957
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.005444
- TAT(Tat/SPI): 0.001518
- LIPO(Sec/SPII): 0.001815
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MSWFDKLFGEDNDSNDDLIHRKKKRRQESQNIDNDHDSLLPQNNDIYSRPRGKFRFPMSVAYENENVEQSADTISDEKEQYHRDYRKQSHDSRSQKRHRRRRNQTTEEQNYSEQRGNSKISQQSIKYKDHSHYHTNKPGTYVSAINGIEKETHKPKTHNMYSNNTNHRAKDSTPDYHKESFKTSEVPSAIFGTMKPKKLENGRIPVSKPSEKVESDKQKYDKYVAKTQTSQNKQLEQEKQNDSVVKQGTASKSSDENVSSTTKSMPNYSKVDNTIKIENIYASQIVEEIRRERERKVLQKRRFKKALQQKREEHKNEEQDAIQRAIDEMYAKQAERYVGDSSLNDDSDLTDNSTDASQLHTNGIENETVSNDENKQASIQNEDTNDTHVDESPYNYEEVSLNQVSTTKQLSDDEVTVSNVTSQHQSALQHNVEVNDKDELKNQSRLIADSEEDGATNKEEYSGSQIDDAEFYELNDTEVDEDTTSNIEDNTNRNASEMHVDAPKTQEYAVTESQVNNIDKTVDNEIELAPRHKKDDQTNLSVNSLKTNDVNDNHVVEDSSMNEIEKNNAEITENVQNEAAESEQNVEEKTIENVNPKKQTEKVSTLSKRPFNVVMTPSDKKRMMDRKKHSKVNVPELKPVQSKQAVSERMPASQATPSSRSDSQESNTNAYKTNNMTSNNVENNQLIGHAETENDYQNAQQYSEQKPSVDSTQTEIFEESQDDNQLENEQVDQSTSSSVSEVSDITEESEETTHPNNTSGQQDNDDQQKDLQSSFSNKNEDTANENRPRTNQQDVATNQAVQTSKPMIRKGPNIKLPSVSLLEEPQVIESDEDWITDKKKELNDALFYFNVPAEVQDVTEGPSVTRFELSVEKGVKVSRITALQDDIKMALAAKDIRIEAPIPGTSRVGIEVPNQNPTTVNLRSIIESPSFKNAESKLTVAMGYRINNEPLLMDIAKTPHALIAGATGSGKSVCINSILMSLLYKNHPEELRLLLIDPKMVELAPYNGLPHLVAPVITDVKAATQSLKWAVEEMERRYKLFAHYHVRNITAFNKKAPYDERMPKIVIVIDELADLMMMAPQEVEQSIARIAQKARACGIHMLVATQRPSVNVITGLIKANIPTRIAFMVSSSVDSRTILDSGGAERLLGYGDMLYLGSGMNKPIRVQGTFVSDDEIDDVVDFIKQQREPDYLFEEKELLKKTQTQSQDELFDDVCAFMVNEGHISTSLIQRHFQIGYNRAARIIDQLEQLGYVSSANGSKPRDVYVTEADLNKE
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: murC < SAOUHSC_01857 < SAOUHSC_01858 < SAOUHSC_01859 < SAOUHSC_01860 < SAOUHSC_01861predicted SigA promoter [3] : SAOUHSC_01854 < SAOUHSC_01855 < murC < SAOUHSC_01857 < SAOUHSC_01858 < SAOUHSC_01859predicted SigA promoter [3] : SAOUHSC_01855 < murC < SAOUHSC_01857 < SAOUHSC_01858 < SAOUHSC_01859 < SAOUHSC_01860 < SAOUHSC_01861
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
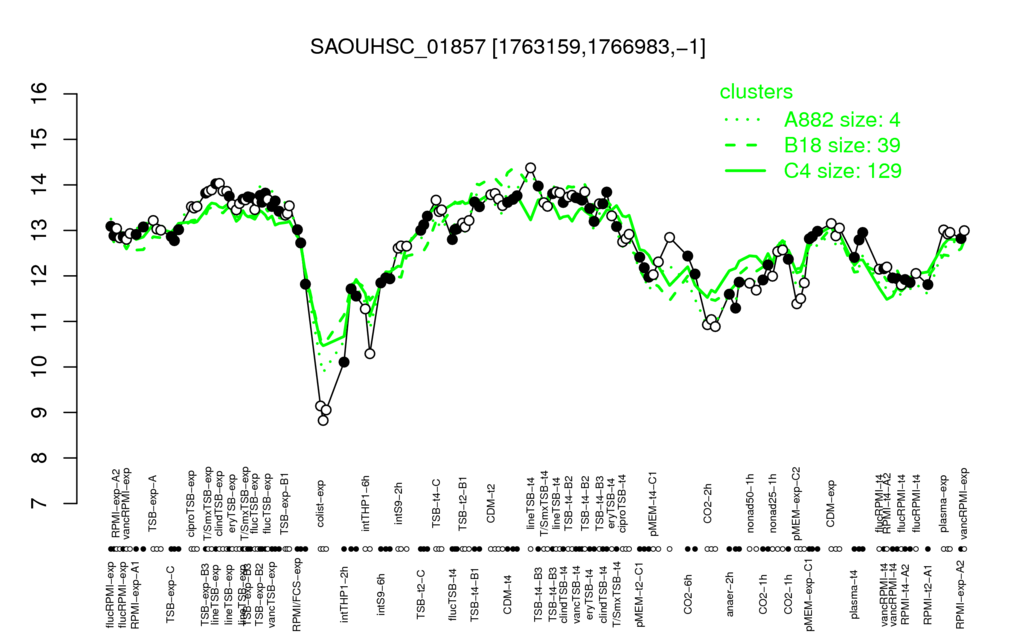 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 3.3 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
