Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01241
- pan locus tag?: SAUPAN003565000
- symbol: polC
- pan gene symbol?: polC
- synonym:
- product: DNA polymerase III PolC
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3920266 NCBI
- RefSeq: YP_499774 NCBI
- BioCyc: G1I0R-1159 BioCyc
- MicrobesOnline: 1289688 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601
3661
3721
3781
3841
3901
3961
4021
4081
4141
4201
4261ATGACAGAGCAACAAAAATTTAAAGTGCTTGCTGATCAAATTAAAATTTCAAATCAATTA
GATGCTGAAATTTTAAATTCAGGTGAACTGACACGTATAGATGTTTCTAACAAAAACAGA
ACATGGGAATTTCATATTACATTACCACAATTCTTAGCTCATGAAGATTATTTATTATTT
ATAAATGCAATAGAGCAAGAGTTTAAAGATATCGCCAACGTTACATGTCGTTTTACGGTA
ACAAATGGCACGAATCAAGATGAACATGCAATTAAATACTTTGGGCACTGTATTGACCAA
ACAGCTTTATCTCCAAAAGTTAAAGGTCAATTGAAACAGAAAAAGCTTATTATGTCTGGA
AAAGTATTAAAAGTAATGGTATCAAATGACATTGAACGTAATCATTTTGATAAGGCATGT
AATGGAAGTCTTATCAAAGCGTTTAGAAATTGTGGTTTTGATATCGATAAAATCATATTC
GAAACAAATGATAATGATCAAGAACAAAACTTAGCTTCTTTAGAAGCACATATTCAAGAA
GAAGACGAACAAAGTGCACGATTGGCAACAGAGAAACTTGAAAAAATGAAAGCTGAAAAA
GCGAAACAACAAGATAACAACGAAAGTGCTGTCGATAAGTGTCAAATTGGTAAGCCGATT
CAAATTGAAAATATTAAACCAATTGAATCTATTATTGAGGAAGAGTTTAAAGTTGCAATA
GAGGGTGTCATTTTTGATATAAACTTAAAAGAACTTAAAAGTGGTCGCCATATCGTAGAA
ATTAAAGTGACTGACTATACGGACTCTTTAGTTTTAAAAATGTTTACTCGTAAAAACAAA
GATGATTTAGAACATTTTAAAGCGCTAAGTGTTGGTAAATGGGTTAGGGCTCAAGGTCGT
ATTGAAGAAGATACATTTATTAGAGATTTAGTTATGATGATGTCTGATATTGAAGAGATT
AAAAAAGCGACAAAAAAAGATAAGGCTGAAGAAAAGCGTGTAGAATTCCACTTGCATACT
GCAATGAGCCAAATGGATGGTATACCCAATATTGGTGCGTATGTTAAACAGGCAGCAGAC
TGGGGACATCCAGCCATTGCGGTTACAGACCATAATGTTGTGCAAGCATTTCCAGATGCT
CACGCAGCAGCGGAAAAACATGGCATTAAAATGATATACGGTATGGAAGGTATGTTAGTT
GATGATGGTGTTCCGATTGCATACAAACCACAAGATGTCGTATTAAAAGATGCTACTTAT
GTTGTGTTCGACGTTGAGACAACTGGTTTATCAAATCAGTATGATAAAATCATCGAGCTT
GCAGCTGTGAAAGTTCATAACGGTGAAATCATCGATAAGTTTGAAAGGTTTAGTAATCCG
CATGAACGATTATCGGAAACGATTATCAATTTGACGCATATTACTGATGATATGTTAGTA
GATGCCCCTGAGATTGAAGAAGTACTTACAGAGTTTAAAGAATGGGTTGGCGATGCGATA
TTCGTAGCGCATAATGCTTCGTTTGATATGGGCTTCATCGATACGGGATATGAACGTCTT
GGGTTTGGACCATCAACGAATGGTGTTATCGATACTTTAGAATTATCTCGTACGATTAAT
ACTGAATATGGTAAACATGGTTTGAATTTCTTGGCTAAAAAATATGGCGTAGAATTAACG
CAACATCACCGTGCCATTTATGATACAGAAGCAACAGCTTACATTTTCATAAAAATGGTT
CAACAAATGAAAGAATTAGGCGTATTAAATCATAACGAAATCAACAAAAAACTCAGTAAT
GAAGATGCATATAAACGTGCAAGACCTAGTCATGTCACATTAATTGTACAAAACCAACAA
GGTCTTAAAAATCTATTTAAAATTGTAAGTGCATCATTGGTGAAGTATTTCTACCGTACA
CCTCGAATTCCACGTTCATTGTTAGATGAATATCGTGAGGGATTATTGGTAGGTACAGCG
TGTGATGAAGGTGAATTATTTACGGCAGTTATGCAGAAGGACCAGAGTCAAGTTGAAAAA
ATTGCCAAATATTATGATTTTATTGAAATTCAACCACCGGCACTTTATCAAGATTTAATT
GATAGAGAGCTTATTAGAGATACTGAAACATTACATGAAATTTATCAACGTTTAATACAT
GCAGGTGACACAGCGGGTATACCTGTTATTGCGACAGGAAATGCACACTATTTGTTTGAA
CATGATGGTATCGCACGTAAAATTTTAATAGCATCACAACCCGGCAATCCACTTAATCGC
TCAACTTTACCGGAAGCACATTTTAGAACTACAGATGAAATGTTAAACGAGTTTCATTTT
TTAGGTGAAGAAAAAGCGCATGAAATTGTTGTGAAAAATACAAACGAATTAGCAGATCGA
ATTGAACGTGTTGTTCCTATTAAAGATGAATTATACACACCGCGTATGGAAGGTGCTAAC
GAAGAAATTAGAGAACTAAGTTATGCAAATGCGCGTAAACTGTATGGTGAAGACCTGCCT
CAAATCGTAATTGATCGATTAGAAAAAGAATTAAAAAGTATTATCGGTAATGGATTTGCG
GTAATTTACTTAATTTCGCAACGTTTAGTTAAAAAATCATTAGATGATGGATACTTAGTT
GGTTCCCGTGGTTCAGTAGGTTCTAGTTTTGTAGCGACAATGACTGAGATTACTGAAGTA
AACCCGTTACCGCCACACTATATTTGTCCGAACTGTAAAACGAGTGAATTTTTCAATGAT
GGTTCAGTAGGATCAGGATTTGATTTACCTGATAAGACGTGTGAAACTTGTGGAGCGCCA
CTTATTAAAGAAGGACAAGATATTCCGTTTGAAACATTTTTAGGATTTAAGGGAGATAAA
GTTCCTGATATCGACTTAAACTTTAGTGGTGAATATCAACCGAATGCCCATAACTACACA
AAAGTATTATTTGGTGAGGATAAAGTATTCCGTGCAGGTACAATTGGTACTGTTGCTGAA
AAGACTGCTTTTGGTTATGTTAAAGGTTATTTGAATGATCAAGGTATCCACAAAAGAGGT
GCTGAAATAGATCGACTCGTTAAAGGATGTACAGGTGTTAAACGTACAACTGGACAGCAT
CCAGGGGGTATTATTGTAGTACCTGATTACATGGATATTTATGATTTTACGCCGATACAA
TATCCTGCCGATGATCAAAATTCAGCATGGATGACGACACATTTTGATTTCCATTCTATT
CATGATAATGTATTAAAACTTGATATACTTGGACACGATGATCCAACAATGATTCGTATG
CTTCAAGATTTATCAGGAATTGATCCAAAAACAATACCTGTAGATGATAAAGAAGTTATG
CAGATATTTAGTACACCTGAAAGTTTGGGTGTTACTGAAGATGAAATTTTATGTAAAACA
GGTACATTTGGGGTACCAGAATTCGGTACAGGATTCGTGCGTCAAATGTTAGAAGATACA
AAGCCAACAACATTTTCTGAATTAGTTCAAATCTCAGGATTATCTCATGGTACAGATGTG
TGGTTAGGCAATGCTCAAGAATTAATTAAAACCGGTATATGTGATTTATCAAGTGTAATT
GGTTGTCGTGATGATATCATGGTTTATTTAATGTATGCTGGTTTAGAACCATCAATGGCT
TTTAAAATAATGGAGTCAGTACGTAAAGGTAAAGGTTTAACTGAAGAAATGATTGAAACG
ATGAAAGAAAATGAAGTGCCAGATTGGTATTTAGATTCATGTCTTAAAATTAAGTACATG
TTCCCTAAAGCCCATGCAGCAGCATACGTTTTAATGGCAGTACGTATCGCATATTTCAAA
GTACATCATCCACTTTATTACTATGCATCTTACTTTACAATTCGTGCGTCAGACTTTGAT
TTAATCACGATGATTAAAGATAAAACAAGCATTCGAAATACTGTAAAAGACATGTATTCT
CGCTATATGGATCTAGGTAAAAAAGAAAAAGACGTATTAACAGTCTTGGAAATTATGAAT
GAAATGGCGCATCGAGGTTATCGAATGCAACCGATTAGTTTAGAAAAGAGTCAGGCGTTC
GAATTTATCATTGAAGGCGATACACTTATTCCGCCGTTCATATCAGTGCCTGGGCTTGGC
GAAAACGTTGCGAAACGAATTGTTGAAGCTCGTGACGATGGCCCATTTTTATCAAAAGAA
GATTTAAACAAAAAAGCTGGATTATCTCAGAAAATTATTGAGTATTTAGATGAGTTAGGC
TCATTACCGAATTTACCAGATAAAGCTCAACTTTCGATATTTGATATGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3660
3720
3780
3840
3900
3960
4020
4080
4140
4200
4260
4311
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01241
- symbol: PolC
- description: DNA polymerase III PolC
- length: 1436
- theoretical pI: 5.0377
- theoretical MW: 162459
- GRAVY: -0.325905
⊟Function[edit | edit source]
- reaction: EC 2.7.7.7? ExPASyDNA-directed DNA polymerase Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1)
- TIGRFAM: DNA metabolism DNA replication, recombination, and repair DNA polymerase III, alpha subunit, Gram-positive type (TIGR01405; EC 2.7.7.7; HMM-score: 2033.4)and 5 moreDNA metabolism DNA replication, recombination, and repair DNA polymerase III, alpha subunit (TIGR00594; EC 2.7.7.7; HMM-score: 217.9)DNA metabolism Degradation of DNA exonuclease, DNA polymerase III, epsilon subunit family (TIGR00573; EC 2.7.7.7; HMM-score: 184.9)DNA metabolism DNA replication, recombination, and repair putative DnaQ family exonuclease/DinG family helicase (TIGR01407; HMM-score: 149.7)DNA metabolism DNA replication, recombination, and repair DNA polymerase III, epsilon subunit (TIGR01406; EC 2.7.7.7; HMM-score: 94.7)Transcription RNA processing ribonuclease T (TIGR01298; EC 3.1.13.-; HMM-score: 45.4)
- TheSEED :
- DNA polymerase III polC-type (EC 2.7.7.7)
- PFAM: NTP_transf (CL0260) DNA_pol3_alpha; Bacterial DNA polymerase III alpha NTPase domain (PF07733; HMM-score: 203.9)and 13 moreRNase_H (CL0219) RNase_T; Exonuclease (PF00929; HMM-score: 161.2)TIM_barrel (CL0036) PHP; PHP domain (PF02811; HMM-score: 113.2)no clan defined DNA_pol3_finger; Bacterial DNA polymerase III alpha subunit finger domain (PF17657; HMM-score: 106.6)DnaA_N (CL0494) DNA_pol3_a_NII; DNA polymerase III polC-type N-terminus II (PF11490; HMM-score: 75.4)no clan defined DNA_pol3_a_NI; DNA polymerase III polC-type N-terminus I (PF14480; HMM-score: 58.8)HHH (CL0198) HHH_6; Helix-hairpin-helix motif (PF14579; HMM-score: 38)HHH_3; Helix-hairpin-helix motif (PF12836; HMM-score: 34.1)OB (CL0021) tRNA_anti-codon; OB-fold nucleic acid binding domain (PF01336; HMM-score: 28.1)RNase_H (CL0219) RNase_H_2; RNase_H superfamily (PF13482; HMM-score: 19)HHH (CL0198) T2SSK; Type II secretion system (T2SS), protein K (PF03934; HMM-score: 17)HHH_5; Helix-hairpin-helix domain (PF14520; HMM-score: 13.6)HTH (CL0123) Ribosomal_S19e; Ribosomal protein S19e (PF01090; HMM-score: 12.8)Zn_Beta_Ribbon (CL0167) Auto_anti-p27; Sjogren's syndrome/scleroderma autoantigen 1 (Autoantigen p27) (PF06677; HMM-score: 12)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9855
- Cytoplasmic Membrane Score: 0.0024
- Cell wall & surface Score: 0.0003
- Extracellular Score: 0.0119
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.000845
- TAT(Tat/SPI): 0.000099
- LIPO(Sec/SPII): 0.000185
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTEQQKFKVLADQIKISNQLDAEILNSGELTRIDVSNKNRTWEFHITLPQFLAHEDYLLFINAIEQEFKDIANVTCRFTVTNGTNQDEHAIKYFGHCIDQTALSPKVKGQLKQKKLIMSGKVLKVMVSNDIERNHFDKACNGSLIKAFRNCGFDIDKIIFETNDNDQEQNLASLEAHIQEEDEQSARLATEKLEKMKAEKAKQQDNNESAVDKCQIGKPIQIENIKPIESIIEEEFKVAIEGVIFDINLKELKSGRHIVEIKVTDYTDSLVLKMFTRKNKDDLEHFKALSVGKWVRAQGRIEEDTFIRDLVMMMSDIEEIKKATKKDKAEEKRVEFHLHTAMSQMDGIPNIGAYVKQAADWGHPAIAVTDHNVVQAFPDAHAAAEKHGIKMIYGMEGMLVDDGVPIAYKPQDVVLKDATYVVFDVETTGLSNQYDKIIELAAVKVHNGEIIDKFERFSNPHERLSETIINLTHITDDMLVDAPEIEEVLTEFKEWVGDAIFVAHNASFDMGFIDTGYERLGFGPSTNGVIDTLELSRTINTEYGKHGLNFLAKKYGVELTQHHRAIYDTEATAYIFIKMVQQMKELGVLNHNEINKKLSNEDAYKRARPSHVTLIVQNQQGLKNLFKIVSASLVKYFYRTPRIPRSLLDEYREGLLVGTACDEGELFTAVMQKDQSQVEKIAKYYDFIEIQPPALYQDLIDRELIRDTETLHEIYQRLIHAGDTAGIPVIATGNAHYLFEHDGIARKILIASQPGNPLNRSTLPEAHFRTTDEMLNEFHFLGEEKAHEIVVKNTNELADRIERVVPIKDELYTPRMEGANEEIRELSYANARKLYGEDLPQIVIDRLEKELKSIIGNGFAVIYLISQRLVKKSLDDGYLVGSRGSVGSSFVATMTEITEVNPLPPHYICPNCKTSEFFNDGSVGSGFDLPDKTCETCGAPLIKEGQDIPFETFLGFKGDKVPDIDLNFSGEYQPNAHNYTKVLFGEDKVFRAGTIGTVAEKTAFGYVKGYLNDQGIHKRGAEIDRLVKGCTGVKRTTGQHPGGIIVVPDYMDIYDFTPIQYPADDQNSAWMTTHFDFHSIHDNVLKLDILGHDDPTMIRMLQDLSGIDPKTIPVDDKEVMQIFSTPESLGVTEDEILCKTGTFGVPEFGTGFVRQMLEDTKPTTFSELVQISGLSHGTDVWLGNAQELIKTGICDLSSVIGCRDDIMVYLMYAGLEPSMAFKIMESVRKGKGLTEEMIETMKENEVPDWYLDSCLKIKYMFPKAHAAAYVLMAVRIAYFKVHHPLYYYASYFTIRASDFDLITMIKDKTSIRNTVKDMYSRYMDLGKKEKDVLTVLEIMNEMAHRGYRMQPISLEKSQAFEFIIEGDTLIPPFISVPGLGENVAKRIVEARDDGPFLSKEDLNKKAGLSQKIIEYLDELGSLPNLPDKAQLSIFDM
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [4] : S508 > rpsB > SAOUHSC_01233 > tsf > S509 > pyrH > frr > S510 > S511 > SAOUHSC_01237 > SAOUHSC_01238 > S512 > SAOUHSC_01239 > SAOUHSC_01240 > S513 > polC > S514 > S515 > SAOUHSC_01242 > nusA > SAOUHSC_01244 > SAOUHSC_01245 > infBpredicted SigA promoter [4] : S513 > polC > S514 > S515 > SAOUHSC_01242 > nusA > SAOUHSC_01244 > SAOUHSC_01245 > infB
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
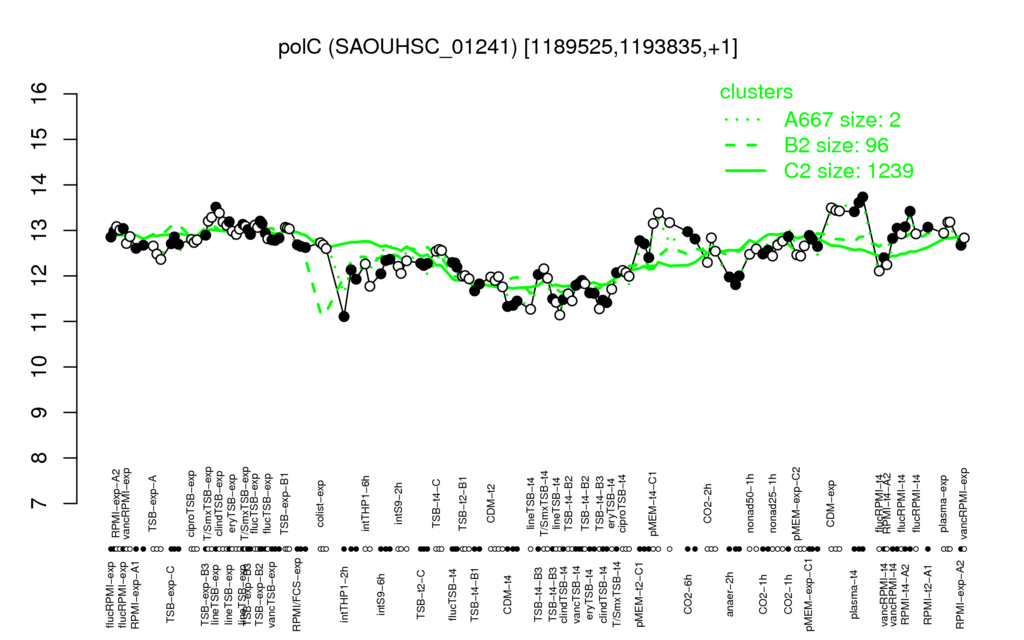 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
R Inoue, C Kaito, M Tanabe, K Kamura, N Akimitsu, K Sekimizu
Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus.
Mol Genet Genomics: 2001, 266(4);564-71
[PubMed:11810227] [WorldCat.org] [DOI] (P p)
