⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00908
- pan locus tag?: SAUPAN003107000
- symbol: SAOUHSC_00908
- pan gene symbol?: cdr
- synonym:
- product: coenzyme A disulfide reductase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00908
- symbol: SAOUHSC_00908
- product: coenzyme A disulfide reductase
- replicon: chromosome
- strand: -
- coordinates: 877458..878774
- length: 1317
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920795 NCBI
- RefSeq: YP_499461 NCBI
- BioCyc: G1I0R-851 BioCyc
- MicrobesOnline: 1289372 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261ATGCCCAAAATAGTCGTAGTCGGAGCAGTCGCTGGCGGTGCAACATGTGCCAGCCAAATT
CGACGTTTAGATAAAGAAAGTGACATTATTATTTTTGAAAAAGATCGTGATATGAGCTTT
GCTAATTGTGCATTGCCTTATGTCATTGGCGAAGTTGTTGAAGATAGAAGATATGCTTTA
GCGTATACACCTGAAAAATTTTATGATAGAAAGCAAATTACAGTAAAAACTTATCATGAA
GTTATTGCAATCAATGATGAAAGACAAACTGTATCTGTATTAAATAGAAAGACAAACGAA
CAATTTGAAGAATCTTACGATAAACTCATTTTAAGCCCTGGTGCAAGTGCAAATAGCCTT
GGCTTTGAAAGTGATATTACATTTACACTTAGAAATTTAGAAGACACTGATGCTATCGAT
CAATTCATCAAAGCAAATCAAGTTGATAAAGTATTGGTTGTAGGTGCAGGTTATGTTTCA
TTAGAAGTTCTTGAAAATCTTTATGAACGTGGTTTACACCCTACTTTAATTCATCGATCT
GATAAGATAAATAAATTAATGGATGCCGACATGAATCAACCTATACTTGATGAATTAGAT
AAGCGGGAGATTCCATACCGTTTAAATGAGGAAATTAATGCTATCAATGGAAATGAAATT
ACATTTAAATCAGGAAAAGTTGAACATTACGATATGATTATTGAAGGTGTCGGTACTCAC
CCCAATTCAAAATTTATCGAAAGTTCAAATATCAAACTTGATCGAAAAGGTTTCATACCG
GTAAACGATAAATTTGAAACAAATGTTCCAAACATTTATGCAATAGGCGATATTGCAACA
TCACATTATCGACATGTCGATCTACCGGCTAGTGTTCCTTTAGCTTGGGGCGCTCACCGT
GCAGCAAGTATTGTTGCCGAACAAATTGCTGGAAATGACACTATTGAATTCAAAGGCTTC
TTAGGCAACAATATTGTGAAGTTCTTTGATTATACATTTGCGAGTGTCGGCGTTAAACCA
AACGAACTAAAGCAATTTGACTATAAAATGGTAGAAGTCACTCAAGGTGCACACGCGAAT
TATTACCCAGGAAATTCCCCTTTACACTTAAGAGTATATTATGACACTTCAAACCGTCAG
ATTTTAAGAGCAGCTGCAGTAGGAAAAGAAGGTGCAGATAAACGTATTGATGTACTATCG
ATGGCAATGATGAACCAGCTAACTGTAGATGAGTTAACTGAGTTTGAAGTGGCTTATGCA
CCACCATATAGCCACCCTAAAGATTTAATCAATATGATTGGTTACAAAGCTAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1317
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00908
- symbol: SAOUHSC_00908
- description: coenzyme A disulfide reductase
- length: 438
- theoretical pI: 5.23597
- theoretical MW: 49289.5
- GRAVY: -0.311416
⊟Function[edit | edit source]
- reaction: EC 1.8.1.14? ExPASyCoA-disulfide reductase 2 CoA + NADP+ = CoA-disulfide + NADPH
- TIGRFAM: Cellular processes Detoxification CoA-disulfide reductase (TIGR03385; EC 1.8.1.14; HMM-score: 687.1)and 20 moreCentral intermediary metabolism Nitrogen metabolism nitrite reductase [NAD(P)H], large subunit (TIGR02374; EC 1.7.1.4; HMM-score: 104.9)dihydrolipoyl dehydrogenase (TIGR01350; EC 1.8.1.4; HMM-score: 104.5)Cellular processes Detoxification mercury(II) reductase (TIGR02053; EC 1.16.1.1; HMM-score: 70.8)Energy metabolism Electron transport thioredoxin-disulfide reductase (TIGR01292; EC 1.8.1.9; HMM-score: 63.5)Unknown function Enzymes of unknown specificity putative bacillithiol system oxidoreductase, YpdA family (TIGR04018; EC 1.8.-.-; HMM-score: 63.3)Energy metabolism Electron transport glutathione-disulfide reductase (TIGR01421; EC 1.8.1.7; HMM-score: 59.4)pyridine nucleotide-disulfide oxidoreductase family protein (TIGR03169; HMM-score: 57.3)trypanothione-disulfide reductase (TIGR01423; EC 1.8.1.12; HMM-score: 54)Energy metabolism Electron transport glutathione-disulfide reductase (TIGR01424; EC 1.8.1.7; HMM-score: 52.6)thioredoxin and glutathione reductase (TIGR01438; EC 1.6.4.-; HMM-score: 48.1)putative alkyl hydroperoxide reductase F subunit (TIGR03143; EC 1.6.4.-; HMM-score: 47.2)mycothione reductase (TIGR03452; EC 1.8.1.15; HMM-score: 42.3)Cellular processes Detoxification alkyl hydroperoxide reductase subunit F (TIGR03140; EC 1.8.1.-; HMM-score: 30.4)Cellular processes Adaptations to atypical conditions alkyl hydroperoxide reductase subunit F (TIGR03140; EC 1.8.1.-; HMM-score: 30.4)Amino acid biosynthesis Glutamate family glutamate synthase (NADPH), homotetrameric (TIGR01316; EC 1.4.1.13; HMM-score: 29.6)putative selenate reductase, YgfK subunit (TIGR03315; HMM-score: 25.6)glutamate synthase, small subunit (TIGR01318; HMM-score: 15.5)glutamate synthase, NADH/NADPH, small subunit (TIGR01317; EC 1.4.1.-; HMM-score: 12.7)Protein synthesis tRNA and rRNA base modification tRNA U-34 5-methylaminomethyl-2-thiouridine biosynthesis protein MnmC, C-terminal domain (TIGR03197; HMM-score: 12.6)Unknown function Enzymes of unknown specificity flavoprotein, HI0933 family (TIGR00275; HMM-score: 11.5)
- TheSEED :
- CoA-disulfide reductase (EC 1.8.1.14)
- PFAM: NADP_Rossmann (CL0063) Pyr_redox_2; Pyridine nucleotide-disulphide oxidoreductase (PF07992; HMM-score: 176.9)and 12 morePyr_redox; Pyridine nucleotide-disulphide oxidoreductase (PF00070; HMM-score: 63.5)Pyr_redox_3; Pyridine nucleotide-disulphide oxidoreductase (PF13738; HMM-score: 49.4)Reductase_C (CL0608) Pyr_redox_dim; Pyridine nucleotide-disulphide oxidoreductase, dimerisation domain (PF02852; HMM-score: 39.1)NADP_Rossmann (CL0063) DAO; FAD dependent oxidoreductase (PF01266; HMM-score: 18.4)NAD_binding_8; NAD(P)-binding Rossmann-like domain (PF13450; HMM-score: 16.2)Lys_Orn_oxgnase; L-lysine 6-monooxygenase/L-ornithine 5-monooxygenase (PF13434; HMM-score: 16.1)NAD_binding_7; Putative NAD(P)-binding (PF13241; HMM-score: 15.8)RNase_H (CL0219) DDE_Tnp_1; Transposase DDE domain (PF01609; HMM-score: 15.5)MurF-HprK_N (CL0365) MurD-like_N; Mur ligase MurD-like, N-terminal domain (PF21799; HMM-score: 13.1)NADP_Rossmann (CL0063) Shikimate_DH; Shikimate / quinate 5-dehydrogenase (PF01488; HMM-score: 13)HI0933_like; HI0933-like protein Rossmann domain (PF03486; HMM-score: 13)FAD_binding_2; FAD binding domain (PF00890; HMM-score: 10.9)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: FAD
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.971
- Cytoplasmic Membrane Score: 0.0252
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0037
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 0.83
- Signal peptide possibility: -0.5
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: Signal peptide LIPO(Sec/SPII) length 15 aa
- SP(Sec/SPI): 0.028234
- TAT(Tat/SPI): 0.001531
- LIPO(Sec/SPII): 0.631807
- Cleavage Site: CS pos: 15-16. GAT-CA. Pr: 0.6133
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MPKIVVVGAVAGGATCASQIRRLDKESDIIIFEKDRDMSFANCALPYVIGEVVEDRRYALAYTPEKFYDRKQITVKTYHEVIAINDERQTVSVLNRKTNEQFEESYDKLILSPGASANSLGFESDITFTLRNLEDTDAIDQFIKANQVDKVLVVGAGYVSLEVLENLYERGLHPTLIHRSDKINKLMDADMNQPILDELDKREIPYRLNEEINAINGNEITFKSGKVEHYDMIIEGVGTHPNSKFIESSNIKLDRKGFIPVNDKFETNVPNIYAIGDIATSHYRHVDLPASVPLAWGAHRAASIVAEQIAGNDTIEFKGFLGNNIVKFFDYTFASVGVKPNELKQFDYKMVEVTQGAHANYYPGNSPLHLRVYYDTSNRQILRAAAVGKEGADKRIDVLSMAMMNQLTVDELTEFEVAYAPPYSHPKDLINMIGYKAK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
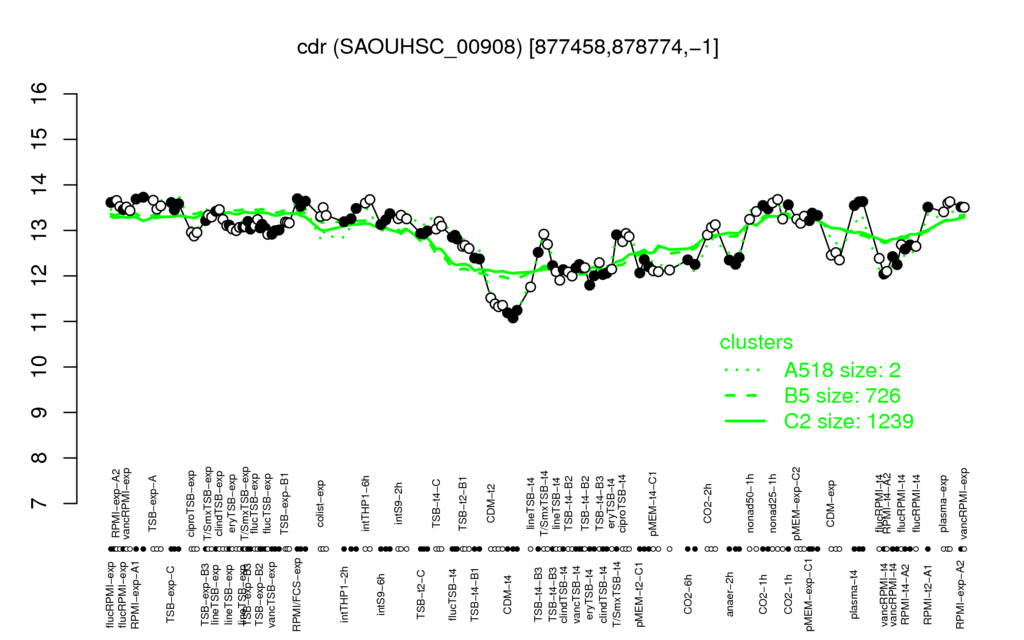 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
J Luba, V Charrier, A Claiborne
Coenzyme A-disulfide reductase from Staphylococcus aureus: evidence for asymmetric behavior on interaction with pyridine nucleotides.
Biochemistry: 1999, 38(9);2725-37
[PubMed:10052943] [WorldCat.org] [DOI] (P p)T Conn Mallett, Jamie R Wallen, P Andrew Karplus, Hiroaki Sakai, Tomitake Tsukihara, Al Claiborne
Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution.
Biochemistry: 2006, 45(38);11278-89
[PubMed:16981688] [WorldCat.org] [DOI] (P p)S B delCardayre, K P Stock, G L Newton, R C Fahey, J E Davies
Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme.
J Biol Chem: 1998, 273(10);5744-51
[PubMed:9488707] [WorldCat.org] [DOI] (P p)S B delCardayre, J E Davies
Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr.
J Biol Chem: 1998, 273(10);5752-7
[PubMed:9488708] [WorldCat.org] [DOI] (P p)
