Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL1224 [new locus tag: SACOL_RS06270 ]
- pan locus tag?: SAUPAN003494000
- symbol: priA
- pan gene symbol?: priA
- synonym:
- product: primosomal protein N`
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL1224 [new locus tag: SACOL_RS06270 ]
- symbol: priA
- product: primosomal protein N`
- replicon: chromosome
- strand: +
- coordinates: 1232873..1235281
- length: 2409
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3237849 NCBI
- RefSeq: YP_186087 NCBI
- BioCyc: see SACOL_RS06270
- MicrobesOnline: 912692 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401ATGATAGCGAAAGTCATAGTCGATGTCGCGTCGAAGAGCGTTGACTATAAATTTGATTAT
ATAATTCCCGAACAACTCGAATCTGTCATCCAACCTGGTGTGCGTGTAATTGTACCTTTT
GGACCAAGAACGATTCAAGGTTATGTAATGGAAGTAACAGCAGAACCTGATGCACAACTT
GACGTTTCGAAGTTAAAAAAAATCATAGAAGTGAAAGATATACAACCAGAATTAACATCA
GAATTAATAGCTTTAAGTGAGTGGATGGGTTCAACTCATGTCATTAAACGTATTTCTATG
CTAGAAGTGATGCTTCCGAGTGCTATTAAAGCGAAGTATAAAAAAGCATTTAAGATGAAA
GATGACATAGAGCTACCTTCAGCTTTATTACAAAAATTTGATAAGCATGGTTACTATTAT
TATAAAGATGCGCAAAAAAATAATGATATTCAATTGCTTATGAAGTTGTTAAAAGATGAT
ATCGTTGAAGAAAAAACGATTCTCACACAAAATATAACTAAAAAAACCAAGCGTGCTGTT
CGTGTCATTGAAGGGTATCATCCTGATGAAGTATTAGCTAAGTTGGAGAAAGTTATTAAA
CAATACGATTTGTATGCTTACTTGTCTGAAGAACAACATAAAACAATATTTTTAACTGAT
ATTGAGGATATGGGCTTTTCAAAATCCAGTTTAGATGGACTTATCAAAAAAGGTTATGTT
GAAAAATATGACGCGGTTGTTGAAAGAGACCCATTTAAAGATCGTGTTTTCGAACAAGAA
TCAAAACAGCAATTAACAGAAGACCAATATAAAGCATATGAAGCGATTAAAGCTAAAATT
GTAAGCCAAGAGCAAGAAACATTTTTACTTCATGGTGTGACGGGATCAGGTAAAACAGAA
GTATATTTACAAACGATAGAAGATGTTTTAAGCCAAGGAAAACAGGCGATGATGTTAGTT
CCTGAAATCGCTCTAACACCGCAAATGGTTTTACGCTTCAAACGTCGATTTGGTGATGAC
GTTGCTGTATTACATTCTGGCTTATCTAATGGGGAACGTTATGATGAATGGCAAAAAATT
AGGGATGGTCGTGCGAGAGTAAGTGTTGGTGCAAGGTCAAGTGTGTTCGCACCTTTCAAA
AATTTAGGGTTAATCATCATTGATGAAGAACATGAATCTACATATAAACAAGAAGATTAT
CCGAGATATCACGCTAGAGAAATTGCCCAATGGCGAAGTGAATATCATCACTGTCCAGTC
ATTTTAGGAAGTGCAACACCATGTCTTGAAAGTTATGCACGAGCTGAAAAAGGCGTTTAT
CATTTGCTATCATTACCAAACAGAGTGAACCAACAAGCTTTACCTGAAATTGATATAGTA
GACATGCGTGAAGAATTGAGTGAAGGTAATCGGTCAATGTTTTCAAAAGATTTACGTGAA
GCCATACAATTAAGATTAGATCGACAGGAACAAGTTGTTTTATTTTTAAATCGACGTGGT
TATGCATCGTTTATGTTATGTCGGGATTGTGGATATGTACCGCAATGTCCAAACTGTGAT
ATTTCATTAACGTATCATAAAACGACAGACTTATTAAAATGTCACTATTGTGGTTACCAA
GAGACGCCACCGAATCAATGTCCAAATTGTGAGAGTGAACACATTCGACAAGTAGGTACT
GGTACTCAGAAAGTTGAAGAACTATTGCAACAAGAATTTGAAGATGCGCGCATAATTAGG
ATGGATGTAGATACAACCTCAAAGAAAGGTGCACATGAAAAGTTATTGACTGAATTCGAA
AAAGGTAACGGTGACATTTTACTAGGTACTCAGATGATTGCGAAAGGATTAGATTATCCA
AATATTACTTTAGTTGGTGTGCTGAATGCAGATACAATGTTAAATTTACCTGATTTTCGG
GCGAGCGAACGTACTTATCAACTATTAACGCAAGTGGCTGGTAGAGCTGGTCGTCATGAA
AAGGCAGGTCAAGTCATCATTCAAACGTATAATCCAGATCATTATTCAATATTGGATGTT
CAAAAAAATGATTATTTAACATTTTATCGTCAGGAAATGGAATATCGTAAATTAGGAAAG
TATCCACCGTATTATTATTTGATTAATTTCACAATCTCACATAAAGAAATGAAGAAGGTT
ATGGTAGCATCGCAGCATGTTCATAAAATTTTATTACAGCATTTAACAGAAAAAGCGCTT
GTACTAGGTCCATCTCCGGCAGCACTTGCGAGAATCAACAATGAATTTAGATTCCAAATT
TTAGTGAAATATAAAAGTGAACCTGGATTATTACAAGCCATTCAGTTTTTAGATGACTAT
TACCATGAAAAATTTATAAAAGAAAAATTAGCATTGAAGATTGATATTGATCCACAGATG
ATGATGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2409
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL1224 [new locus tag: SACOL_RS06270 ]
- symbol: PriA
- description: primosomal protein N`
- length: 802
- theoretical pI: 6.74374
- theoretical MW: 92521.8
- GRAVY: -0.421072
⊟Function[edit | edit source]
- reaction: EC 3.6.4.-? ExPASy
- TIGRFAM: DNA metabolism DNA replication, recombination, and repair primosomal protein N' (TIGR00595; EC 3.6.1.-; HMM-score: 712.9)and 8 moreDNA metabolism DNA replication, recombination, and repair ATP-dependent DNA helicase RecG (TIGR00643; EC 3.6.4.12; HMM-score: 79.2)DNA metabolism DNA replication, recombination, and repair transcription-repair coupling factor (TIGR00580; EC 3.6.1.-; HMM-score: 70.8)DNA metabolism DNA replication, recombination, and repair excinuclease ABC subunit B (TIGR00631; EC 3.1.25.-; HMM-score: 49.8)type IV conjugative transfer system coupling protein TraD (TIGR02759; HMM-score: 14.4)Protein synthesis tRNA and rRNA base modification tRNA 2-selenouridine synthase (TIGR03167; EC 2.9.1.-; HMM-score: 13.9)KaiC domain protein, AF_0351 family (TIGR03880; HMM-score: 12.4)Unknown function General putative alkylphosphonate utilization operon protein PhnA (TIGR00686; HMM-score: 12.1)Purines, pyrimidines, nucleosides, and nucleotides 2'-Deoxyribonucleotide metabolism anaerobic ribonucleoside-triphosphate reductase (TIGR02487; EC 1.17.4.2; HMM-score: 11.6)
- TheSEED :
- Helicase PriA essential for oriC/DnaA-independent DNA replication
- PFAM: no clan defined PriA_3primeBD; 3'DNA-binding domain (3'BD) (PF17764; HMM-score: 80.1)PriA_C; Primosomal protein N C-terminal domain (PF18074; HMM-score: 77)P-loop_NTPase (CL0023) DEAD; DEAD/DEAH box helicase (PF00270; HMM-score: 71)ResIII; Type III restriction enzyme, res subunit (PF04851; HMM-score: 71)and 15 moreHelicase_C; Helicase conserved C-terminal domain (PF00271; HMM-score: 49)Zn_Beta_Ribbon (CL0167) Zn_ribbon_PriA; PriA DNA helicase Cys-rich region (CRR) domain (PF18319; HMM-score: 47.7)P-loop_NTPase (CL0023) Cas3-like_C_2; CRISPR-associated nuclease/helicase Cas3, C-terminal (PF22590; HMM-score: 20.4)PIF1; PIF1-like helicase (PF05970; HMM-score: 18.8)HTH (CL0123) SelB_WHD3; Elongation factor SelB, third winged-helix domain (PF21580; HMM-score: 16.9)P-loop_NTPase (CL0023) Helicase_C_2; Helicase C-terminal domain (PF13307; HMM-score: 15.9)AAA_19; AAA domain (PF13245; HMM-score: 14)AAA_30; AAA domain (PF13604; HMM-score: 13.9)Zn_Beta_Ribbon (CL0167) Zn_ribbon_IS1595; Transposase zinc-ribbon domain (PF12760; HMM-score: 13.1)P-loop_NTPase (CL0023) T2SSE; Type II/IV secretion system protein (PF00437; HMM-score: 13)TrwB_AAD_bind; Type IV secretion-system coupling protein DNA-binding domain (PF10412; HMM-score: 12.9)Zn_Beta_Ribbon (CL0167) Zn_ribbon_6; Double zinc ribbon (PF07191; HMM-score: 11.7)Zn_Ribbon_TF; TFIIB zinc-binding (PF08271; HMM-score: 8.1)Zn_Ribbon_YjdM; PhnA Zinc-Ribbon (PF08274; HMM-score: 7.2)Zn_ribbon_PF0610; PF0610 rubredoxin-like zinc beta-ribbon domain (PF23470; HMM-score: 6.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9806
- Cytoplasmic Membrane Score: 0.0157
- Cell wall & surface Score: 0.0019
- Extracellular Score: 0.0018
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.005634
- TAT(Tat/SPI): 0.00011
- LIPO(Sec/SPII): 0.000239
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIAKVIVDVASKSVDYKFDYIIPEQLESVIQPGVRVIVPFGPRTIQGYVMEVTAEPDAQLDVSKLKKIIEVKDIQPELTSELIALSEWMGSTHVIKRISMLEVMLPSAIKAKYKKAFKMKDDIELPSALLQKFDKHGYYYYKDAQKNNDIQLLMKLLKDDIVEEKTILTQNITKKTKRAVRVIEGYHPDEVLAKLEKVIKQYDLYAYLSEEQHKTIFLTDIEDMGFSKSSLDGLIKKGYVEKYDAVVERDPFKDRVFEQESKQQLTEDQYKAYEAIKAKIVSQEQETFLLHGVTGSGKTEVYLQTIEDVLSQGKQAMMLVPEIALTPQMVLRFKRRFGDDVAVLHSGLSNGERYDEWQKIRDGRARVSVGARSSVFAPFKNLGLIIIDEEHESTYKQEDYPRYHAREIAQWRSEYHHCPVILGSATPCLESYARAEKGVYHLLSLPNRVNQQALPEIDIVDMREELSEGNRSMFSKDLREAIQLRLDRQEQVVLFLNRRGYASFMLCRDCGYVPQCPNCDISLTYHKTTDLLKCHYCGYQETPPNQCPNCESEHIRQVGTGTQKVEELLQQEFEDARIIRMDVDTTSKKGAHEKLLTEFEKGNGDILLGTQMIAKGLDYPNITLVGVLNADTMLNLPDFRASERTYQLLTQVAGRAGRHEKAGQVIIQTYNPDHYSILDVQKNDYLTFYRQEMEYRKLGKYPPYYYLINFTISHKEMKKVMVASQHVHKILLQHLTEKALVLGPSPAALARINNEFRFQILVKYKSEPGLLQAIQFLDDYYHEKFIKEKLALKIDIDPQMMM
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas
- protein localization: Cytoplasmic [1] [2]
- quantitative data / protein copy number per cell: 196 [3]
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
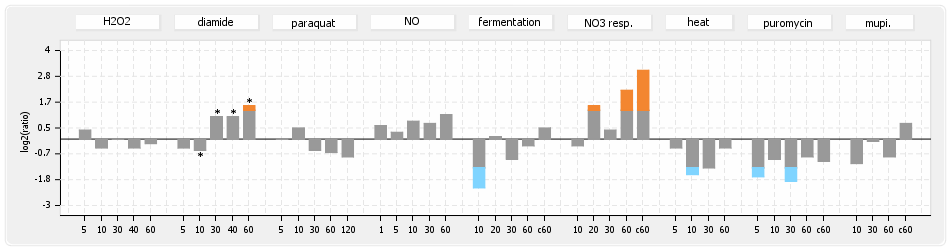
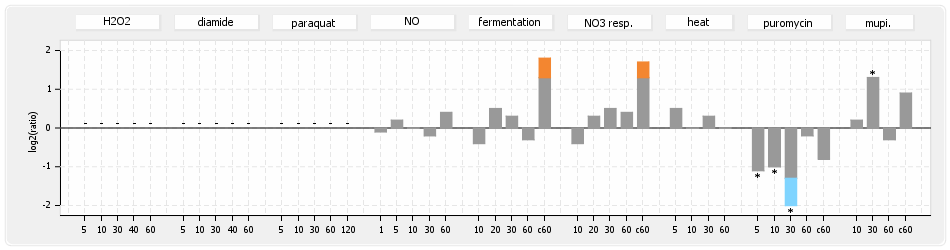
- S.aureus Expression Data Browser: data available for NCTC8325
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e)