Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02363
- pan locus tag?: SAUPAN005414000
- symbol: SAOUHSC_02363
- pan gene symbol?: —
- synonym:
- product: aldehyde dehydrogenase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02363
- symbol: SAOUHSC_02363
- product: aldehyde dehydrogenase
- replicon: chromosome
- strand: -
- coordinates: 2184537..2185964
- length: 1428
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919406 NCBI
- RefSeq: YP_500839 NCBI
- BioCyc: G1I0R-2232 BioCyc
- MicrobesOnline: 1290800 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381ATGAGAGACTACACAAAGCAATACATTAATGGCGAATGGGTAGAAAGTAATAGTAATGAA
ACGATAGAAGTTATAAATCCAGCAACCGAAGAAGTAATCGGGAAAGTTGCTAAAGGTAAT
AAAGCTGATGTTGATAAAGCCGTCGAGGCGGCAGACGATGTTTATTTAGAGTTCCGTCAT
ACATCTGTGAAAGAAAGACAAGCGTTATTAGATAAAATTGTAAAAGAATATGAAAACAGA
AAAGACGATATTGTACAAGCTATTACGGATGAATTAGGTGCTCCTTTATCATTATCTGAG
CGTGTCCATTATCAAATGGGACTAAACCATTTTGTTGCAGCGAGAGACGCATTAGATAAC
TACGAATTTGAAGAACGCCGCGGAGATGATTTAGTTGTTAAAGAAGCAATCGGTGTATCT
GGATTAATTACACCGTGGAACTTCCCTACAAACCAAACATCATTAAAATTAGCAGCAGCA
TTTGCGGCTGGTAGTCCAGTTGTACTTAAACCATCTGAAGAAACACCATTTGCAGCTGTT
ATTTTAGCTGAGATTTTTGATAAAGTCGGTGTTCCTAAAGGTGTATTTAACCTTGTTAAT
GGTGATGGTGCTGGTGTTGGGAATCCTTTATCTGAACATCCTAAAGTACGCATGATGTCA
TTTACAGGATCAGGCCCTACTGGTTCTAAAATTATGGAAAAAGCCGCTAAAGATTTTAAA
AAGGTATCATTAGAGCTTGGTGGCAAATCACCATATATCGTCCTAGATGACGTAGATATT
AAAGAAGCGGCTAAAGCAACAACAGGCAAAGTTGTTAATAATACTGGTCAAGTATGTACA
GCTGGTACACGTGTTTTAGTGCCTAACAAAATTAAAGATGCATTCTTAGCTGAATTAAAA
GAACAATTTAGCCAAGTGCGTGTCGGTAATCCAAGAGAAGATGGTACACAAGTAGGCCCT
ATCATTAGTAAAAAACAATTTGATCAAGTACAAAATTATATTAATAAAGGTATTGAAGAA
GGTGCTGAATTATTTTATGGTGGTCCTGGTAAACCAGAAGGACTTGAAAAAGGATACTTT
GCACGTCCGACAATTTTTATTAATGTAGATAATCAAATGACGATAGCACAAGAAGAAATT
TTTGGGCCAGTAATGTCAGTTATCACTTATAACGATTTAGATGAAGCGATTCAAATTGCA
AATGATACAAAATATGGTTTGGCAGGATATGTTATTGGTAAGGACAAAGAAACATTGCAT
AAAGTAGCTCGTTCTATTGAAGCAGGTACAGTAGAAATAAACGAAGCAGGTAGAAAGCCA
GATTTACCATTTGGTGGCTATAAACAATCTGGTTTAGGTCGTGAATGGGGCGATTATGGT
ATTGAAGAGTTCTTAGAAGTGAAATCTATAGCTGGATATTTTAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1428
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02363
- symbol: SAOUHSC_02363
- description: aldehyde dehydrogenase
- length: 475
- theoretical pI: 4.76962
- theoretical MW: 51968.3
- GRAVY: -0.350526
⊟Function[edit | edit source]
- ⊞reaction: EC 1.2.1.3? ExPASyEC 1.2.1.16? ExPASy
- ⊞TIGRFAM: Cellular processes Adaptations to atypical conditions betaine-aldehyde dehydrogenase (TIGR01804; EC 1.2.1.8; HMM-score: 460)5-carboxymethyl-2-hydroxymuconate semialdehyde dehydrogenase (TIGR02299; EC 1.2.1.60; HMM-score: 449.2)Unknown function Enzymes of unknown specificity aldehyde dehydrogenase, Rv0768 family (TIGR04284; EC 1.2.1.-; HMM-score: 405)Central intermediary metabolism Other succinate-semialdehyde dehydrogenase (TIGR01780; EC 1.2.1.-; HMM-score: 403.8)Energy metabolism Other 2-hydroxymuconic semialdehyde dehydrogenase (TIGR03216; EC 1.2.1.-; HMM-score: 375.8)and 10 more
- ⊞TheSEED :
- Aldehyde dehydrogenase (NAD(+)) (EC 1.2.1.3)
Carbohydrates Central carbohydrate metabolism Methylglyoxal Metabolism Aldehyde dehydrogenase (EC 1.2.1.3)and 2 more - PFAM: ALDH-like (CL0099) Aldedh; Aldehyde dehydrogenase family (PF00171; HMM-score: 563.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MRDYTKQYINGEWVESNSNETIEVINPATEEVIGKVAKGNKADVDKAVEAADDVYLEFRHTSVKERQALLDKIVKEYENRKDDIVQAITDELGAPLSLSERVHYQMGLNHFVAARDALDNYEFEERRGDDLVVKEAIGVSGLITPWNFPTNQTSLKLAAAFAAGSPVVLKPSEETPFAAVILAEIFDKVGVPKGVFNLVNGDGAGVGNPLSEHPKVRMMSFTGSGPTGSKIMEKAAKDFKKVSLELGGKSPYIVLDDVDIKEAAKATTGKVVNNTGQVCTAGTRVLVPNKIKDAFLAELKEQFSQVRVGNPREDGTQVGPIISKKQFDQVQNYINKGIEEGAELFYGGPGKPEGLEKGYFARPTIFINVDNQMTIAQEEIFGPVMSVITYNDLDEAIQIANDTKYGLAGYVIGKDKETLHKVARSIEAGTVEINEAGRKPDLPFGGYKQSGLGREWGDYGIEEFLEVKSIAGYFK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- ⊟interaction partners:
SAOUHSC_00529 elongation factor G [3] (data from MRSA252) SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_01806 pyruvate kinase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [5]
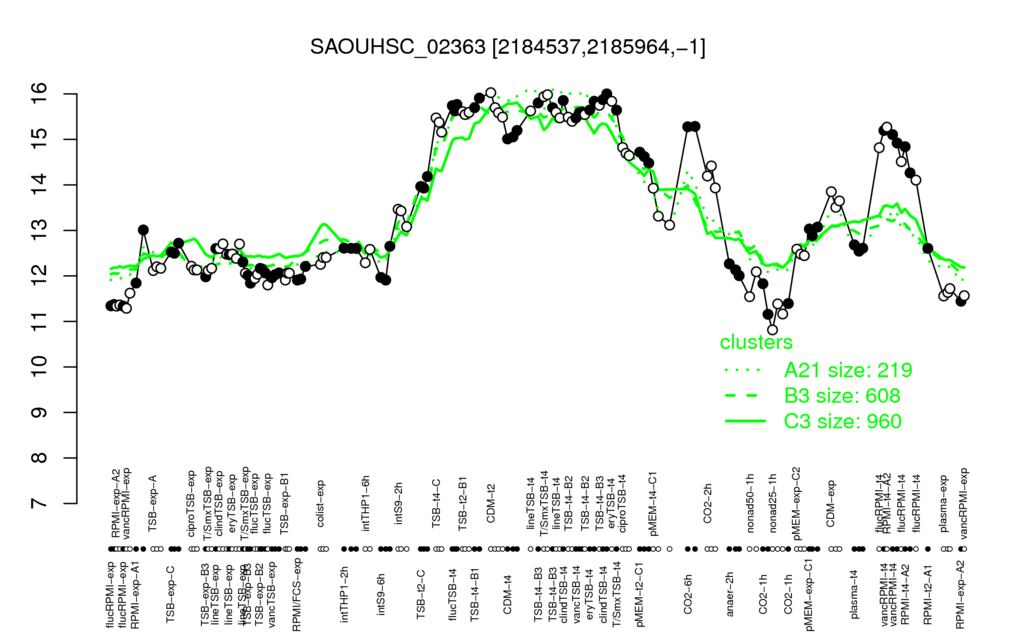 ⊟Multi-gene expression profiles
⊟Multi-gene expression profiles
Click on any data point to display a description of the corresponding condition!
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊞Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊞Other Information[edit | edit source]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Jump up to: 3.0 3.1 3.2 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Markus Bischoff, Paul Dunman, Jan Kormanec, Daphne Macapagal, Ellen Murphy, William Mounts, Brigitte Berger-Bächi, Steven Projan
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon.
J Bacteriol: 2004, 186(13);4085-99
[PubMed:15205410] [WorldCat.org] [DOI] (P p) - ↑ Jump up to: 5.0 5.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
