Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02417
- pan locus tag?: SAUPAN005553000
- symbol: SAOUHSC_02417
- pan gene symbol?: salA
- synonym:
- product:
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3919628 NCBI
- RefSeq:
- BioCyc: G1I0R-2289 BioCyc
- MicrobesOnline: 1290860 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021TTGTTAACGGTAGATCAAGTGAAAGAATTGGTAGGAGAAATTAAAGATCCTATTATAGAT
GTGCCTTTAAAAGAAACAGAAGGTATTGTTGAAGTTTCTATTAAGGAAGAAAAAGAACAT
GTGAGTGTTAAATTAGCAATGGCACAATTAGGTGGTGCACCGCAATTAGATTTACAGATG
GCTGTTGTTAATGCATTAAAAGAAAACGGTGCGAAAACGGTCGGTATACGATTTGAAACA
TTGCCGGAAGAAAAAGTAAATCAATTTAAACCAAAAGAAGAAAATAAACCTAAAACGATA
GAAGGCCTATTATCTCAAAATAATCCAGTCGAATTTATTGCAATAGCCTCCGGTAAAGGT
GGTGTCGGTAAATCTACTGTTGCAGTAAATTTAGCCGTTGCCTTAGCTCGTGAAGGGAAA
AAAGTCGGATTAGTAGATGCCGATATATATGGATTTAGTGTACCAGATATGATGGGTATT
GATGAAAAGCCTGGAATTAAAGGGAAGGAAGTAATTCCAGTTGAACGTCATGGCGTTAAA
GTTATATCAATGGCCTTTTTTGTGGAAGAAAATGCGCCAGTTATATGGAGAGGGCCAATG
TTAGGTAAAATGTTGACGAATTTCTTTACAGAAGTTAAATGGGGAGACATTGAATATTTA
ATACTCGATCTTCCACCTGGAACAGGAGATGTAGCTTTAGATGTTCATACGATGTTACCT
TCAAGTAAGGAAATTATTGTAACGACACCTCATCCTACAGCAGCATTTGTTGCAGCTCGC
GCAGGTGCGATGGCAAAACATACGGATCATTCTATTCTTGGCGTAATTGAAAACATGAGT
TATTTTGAAAGTAAAGAGACGGGTAATAAAGAATATGTCTTTGGCAAAGGTGGCGGTACT
AAGTTAGCTGATGAACTTAATACTCAATTACTTGGGGAATTACCTTTAGAGCAACCATCT
TGGAATCCAAAAGATTTTGCACCGTCAATATATCAATCAGATGATCGTCTAGGTAAAATT
TATAGCTCGATTGCACAAAAAGTTATAGCTGCTACAAATAAATAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1065
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02417
- symbol: SAOUHSC_02417
- description:
- length: 354
- theoretical pI: 5.069
- theoretical MW: 38358
- GRAVY: -0.121751
⊟Function[edit | edit source]
- TIGRFAM: cell division ATPase MinD (TIGR01969; HMM-score: 69.8)and 20 moreCellular processes Cell division septum site-determining protein MinD (TIGR01968; HMM-score: 51.2)helicase/secretion neighborhood CpaE-like protein (TIGR03815; HMM-score: 44.9)Transport and binding proteins Carbohydrates, organic alcohols, and acids capsular exopolysaccharide family (TIGR01007; HMM-score: 36.8)exopolysaccharide/PEP-CTERM locus tyrosine autokinase (TIGR03018; EC 2.7.10.2; HMM-score: 35.3)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides cellulose synthase operon protein YhjQ (TIGR03371; HMM-score: 29.9)arsenical pump-driving ATPase (TIGR04291; EC 3.6.1.-; HMM-score: 28.1)Biosynthesis of cofactors, prosthetic groups, and carriers Chlorophyll and bacteriochlorphyll light-independent protochlorophyllide reductase, iron-sulfur ATP-binding protein (TIGR01281; EC 1.3.7.7; HMM-score: 27.8)Hypothetical proteins Conserved transport-energizing ATPase, TRC40/GET3/ArsA family (TIGR00345; EC 3.6.1.-; HMM-score: 25.7)Transport and binding proteins Carbohydrates, organic alcohols, and acids exopolysaccharide transport protein family (TIGR01005; HMM-score: 25.7)Central intermediary metabolism Nitrogen fixation nitrogenase iron protein (TIGR01287; EC 1.18.6.1; HMM-score: 23.8)chain length determinant protein tyrosine kinase EpsG (TIGR03029; HMM-score: 21.5)Mobile and extrachromosomal element functions Plasmid functions plasmid partitioning protein RepA (TIGR03453; HMM-score: 17.9)DNA repair and recombination protein RadB (TIGR02237; HMM-score: 16.4)Energy metabolism Photosynthesis chlorophyllide reductase iron protein subunit X (TIGR02016; HMM-score: 16.2)Transport and binding proteins Amino acids, peptides and amines LAO/AO transport system ATPase (TIGR00750; EC 2.7.-.-; HMM-score: 15.9)Regulatory functions Protein interactions LAO/AO transport system ATPase (TIGR00750; EC 2.7.-.-; HMM-score: 15.9)Cellular processes Chemotaxis and motility flagellar biosynthesis protein FlhF (TIGR03499; HMM-score: 15)Protein fate Protein and peptide secretion and trafficking signal recognition particle-docking protein FtsY (TIGR00064; HMM-score: 14.1)Central intermediary metabolism Sulfur metabolism adenylyl-sulfate kinase (TIGR00455; EC 2.7.1.25; HMM-score: 12.7)Biosynthesis of cofactors, prosthetic groups, and carriers Heme, porphyrin, and cobalamin putative heme d1 biosynthesis radical SAM protein NirJ2 (TIGR04055; EC 1.3.99.-; HMM-score: 12.6)
- TheSEED :
- Scaffold protein for [4Fe-4S] cluster assembly ApbC, MRP-like
- PFAM: P-loop_NTPase (CL0023) ParA; NUBPL iron-transfer P-loop NTPase (PF10609; HMM-score: 293.2)and 17 moreCbiA; CobQ/CobB/MinD/ParA nucleotide binding domain (PF01656; HMM-score: 51)AAA_31; AAA domain (PF13614; HMM-score: 46.4)MipZ; ATPase MipZ (PF09140; HMM-score: 32.1)Fer4_NifH; 4Fe-4S iron sulfur cluster binding proteins, NifH/frxC family (PF00142; HMM-score: 28.5)ArsA_ATPase; Anion-transporting ATPase (PF02374; HMM-score: 26.3)AAA_25; AAA domain (PF13481; HMM-score: 22.1)VirC1; VirC1 protein (PF07015; HMM-score: 20.4)CLP1_P; mRNA cleavage and polyadenylation factor CLP1 P-loop (PF16575; HMM-score: 19.2)SRP54; SRP54-type protein, GTPase domain (PF00448; HMM-score: 16.5)CBP_BcsQ; Cellulose biosynthesis protein BcsQ (PF06564; HMM-score: 15.9)MeaB; Methylmalonyl Co-A mutase-associated GTPase MeaB (PF03308; HMM-score: 15)nSTAND3; Novel STAND NTPase 3 (PF20720; HMM-score: 13.9)APS_kinase; Adenylylsulphate kinase (PF01583; HMM-score: 13.5)NTPase_1; NTPase (PF03266; HMM-score: 13)NB-ARC; NB-ARC domain (PF00931; HMM-score: 12.1)FTHFS; Formate--tetrahydrofolate ligase (PF01268; HMM-score: 11.5)KTI12; Chromatin associated protein KTI12 (PF08433; HMM-score: 11.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9086
- Cytoplasmic Membrane Score: 0.0357
- Cell wall & surface Score: 0.0004
- Extracellular Score: 0.0554
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003818
- TAT(Tat/SPI): 0.000508
- LIPO(Sec/SPII): 0.000224
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLTVDQVKELVGEIKDPIIDVPLKETEGIVEVSIKEEKEHVSVKLAMAQLGGAPQLDLQMAVVNALKENGAKTVGIRFETLPEEKVNQFKPKEENKPKTIEGLLSQNNPVEFIAIASGKGGVGKSTVAVNLAVALAREGKKVGLVDADIYGFSVPDMMGIDEKPGIKGKEVIPVERHGVKVISMAFFVEENAPVIWRGPMLGKMLTNFFTEVKWGDIEYLILDLPPGTGDVALDVHTMLPSSKEIIVTTPHPTAAFVAARAGAMAKHTDHSILGVIENMSYFESKETGNKEYVFGKGGGTKLADELNTQLLGELPLEQPSWNPKDFAPSIYQSDDRLGKIYSSIAQKVIAATNK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [4] : SAOUHSC_02417 < S940
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
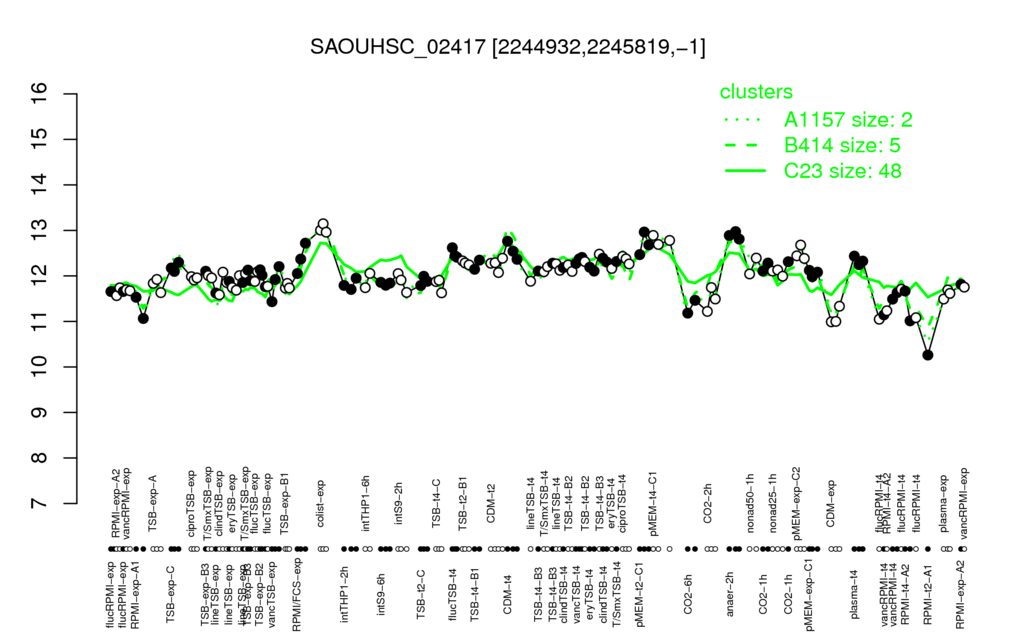 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Anne Berscheid, Peter Sass, Konstantin Weber-Lassalle, Ambrose L Cheung, Gabriele Bierbaum
Revisiting the genomes of the Staphylococcus aureus strains NCTC 8325 and RN4220.
Int J Med Microbiol: 2012, 302(2);84-7
[PubMed:22417616] [WorldCat.org] [DOI] (I p) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
