Jump to navigation
Jump to search
PangenomeCOLN315NCTC8325NewmanUSA300_FPR3757JSNZ04-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159LGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01563
- pan locus tag?: SAUPAN001439000
- symbol: SAOUHSC_01563
- pan gene symbol?: —
- synonym:
- product: phage encoded DNA polymerase I
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01563
- symbol: SAOUHSC_01563
- product: phage encoded DNA polymerase I
- replicon: chromosome
- strand: -
- coordinates: 1497709..1499661
- length: 1953
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3920158 NCBI
- RefSeq: YP_500079 NCBI
- BioCyc: G1I0R-1452 BioCyc
- MicrobesOnline: 1289993 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921ATGAATATAGATATTGAAACATACAGCAGTAACGATATTTCGAAATGTGGTGCCTATAAA
TACACAGAAGCTGAAGATTTCGAAATTTTAATTATAGCTTATTCGATAGATGGTGGAGCG
ATTAGTGCGATTGACATGACTAAAGTAGATAATGAGCCTTTCCACGCTGATTATGAGACG
TTTAAAATTGCTCTATTTGACCCTGCTGTAAAAAAGTATGCATTCAATGCTAATTTCGAA
AGAACTTGTCTTGCTAAACATTTTAATAAACAGATGCCACCTGAAGAATGGATTTGCACA
ATGGTTAATTCAATGCGTATTGGCTTACCTGCTTCGCTTGATAAAGTTGGAGAAGTTTTA
AGACTACAAAGCCAAAAAGATAAAGCAGGTAAAAATTTAATTCGTTATTTCTCTATACCT
TGTAAACCAACAAAAGTTAATGGAGGAAGAACAAGAAACCTACCTGAACATGATCTTGAA
AAATGGCAACAATTTATAGATTACTGTATTCGAGATGTAGAAGTAGAAATGGCGATTGCT
AATAAAATTAAAGACTTTCCAGTAACTGCAATTGAACAAACATATTGGGTTTTTGACCAA
CATATAAACGACAGAGGTATTAAGCTTTCTAAATCATTGATGTTAGGAGCTAATGTGCTC
GATAAGCAGAGTAAAGAAGAATTGCTTAAACAAGCTAAACATATAACAGGTTTAGAAAAT
CCTAATAGTCCTACACAGTTATTGGCTTGGTTAAAGGATGAACAAGGATTAGATATACCT
AATTTACAAAAGAAAACGGTTCAGGAGTACTTAAAAGAAGCAACAGGAAAAGCTAAAAAA
ATGCTAGAAATTAGATTGCAAATGTCTAAAACCAGTGTGAAAAAATACAACAAAATGCAT
GACATGATGTGCAGTGATGAACGGGTAAGAGGTCTGTTTCAATTTTACGGTGCCGGTACT
GGAAGATGGGCAGGTAGAGGTGTACAACTTCAGAATTTAACAAAGCATTATATTTCAGAT
ACTGAATTAGAAATAGCAAGAGATCTTATTAAAGAACAACGTTTTGATGATTTAGATTTA
TTACTCAATGTTCATCCTCAAGACTTATTAAGTCAATTAGTTAGGACGACATTTACTGCT
GAAGAAGGTAATGAACTAGCAGTAAGTGATTTTTCTGCAATAGAGGCAAGAGTCATAGCA
TGGTATGCAAAAGAACAATGGCGTTTAGATGTATTCAACACACACGGAAAGATATATGAA
GCATCGGCTTCTCAAATGTTTAATGTACCGGTAGAAAGCATAACTAAAGGCGACCCTCTC
AGACAAAAAGGAAAAGTGTCCGAATTAGCTTTAGGCTATCAAGGTGGCGCTGGAGCTTTA
AAAGCAATGGGTGCATTGGAAATGGGCATTGAAGAAAACGAGTTACAAGGTTTAGTTGAT
AGTTGGCGTAACGCAAATCCTAACATAGTTAATTTTTGGAAGGCTTGCCAAGAGGCTGCA
ATTAATACTGTAAAATCCCGAAAGACGCATCATACACATGGACTTAGATTTTATATGAAA
AAAGGTTTTCTAATGATTGAACTGCCTAGTGGAAGAGCTTTAGCTTATCCAAAAGCTTTA
GTTGGTGAAAATAGTTGGGGTAGTCAAGTTGTTGAATTTATGGGGTTAGATCTTAACCGT
AAATGGTCAAAGTTAAAAACGTATGGTGGGAAGTTAGTCGAGAATATTGTTCAAGCAACT
GCAAGGGATTTACTTGCGATTTCTATAGCAAGGCTTGAAGCATTAGGTTTTAAAATAGTT
GGCCATGTCCATGATGAAGTAATTGTAGAAATACCTAGAGGTTCAAATGGACTTAAGGAA
ATCGAAACTATCATGAATAAGCCTGTTGATTGGGCAAAAGGATTGAATTTGAATAGTGAC
GGGTTTACTTCTCCGTTTTATATGAAGGATTAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1953
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01563
- symbol: SAOUHSC_01563
- description: phage encoded DNA polymerase I
- length: 650
- theoretical pI: 7.39229
- theoretical MW: 73568.9
- GRAVY: -0.38
⊟Function[edit | edit source]
- reaction: EC 2.7.7.7? ExPASyDNA-directed DNA polymerase Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1)
- TIGRFAM: DNA metabolism DNA replication, recombination, and repair DNA polymerase I (TIGR00593; EC 2.7.7.7; HMM-score: 54.4)
- TheSEED :
- Phage DNA polymerase I (EC 2.7.7.7)
and 1 more - PFAM: no clan defined DNA_pol_A; DNA polymerase family A (PF00476; HMM-score: 87.8)and 2 moreRNase_H (CL0219) DNA_pol_A_exo1; 3'-5' exonuclease (PF01612; HMM-score: 14.3)Beta_propeller (CL0186) KNTC1_N; KNTC1, N-terminal (PF24506; HMM-score: 12)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9075
- Cytoplasmic Membrane Score: 0.0024
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0901
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.009746
- TAT(Tat/SPI): 0.000639
- LIPO(Sec/SPII): 0.001899
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNIDIETYSSNDISKCGAYKYTEAEDFEILIIAYSIDGGAISAIDMTKVDNEPFHADYETFKIALFDPAVKKYAFNANFERTCLAKHFNKQMPPEEWICTMVNSMRIGLPASLDKVGEVLRLQSQKDKAGKNLIRYFSIPCKPTKVNGGRTRNLPEHDLEKWQQFIDYCIRDVEVEMAIANKIKDFPVTAIEQTYWVFDQHINDRGIKLSKSLMLGANVLDKQSKEELLKQAKHITGLENPNSPTQLLAWLKDEQGLDIPNLQKKTVQEYLKEATGKAKKMLEIRLQMSKTSVKKYNKMHDMMCSDERVRGLFQFYGAGTGRWAGRGVQLQNLTKHYISDTELEIARDLIKEQRFDDLDLLLNVHPQDLLSQLVRTTFTAEEGNELAVSDFSAIEARVIAWYAKEQWRLDVFNTHGKIYEASASQMFNVPVESITKGDPLRQKGKVSELALGYQGGAGALKAMGALEMGIEENELQGLVDSWRNANPNIVNFWKACQEAAINTVKSRKTHHTHGLRFYMKKGFLMIELPSGRALAYPKALVGENSWGSQVVEFMGLDLNRKWSKLKTYGGKLVENIVQATARDLLAISIARLEALGFKIVGHVHDEVIVEIPRGSNGLKEIETIMNKPVDWAKGLNLNSDGFTSPFYMKD
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
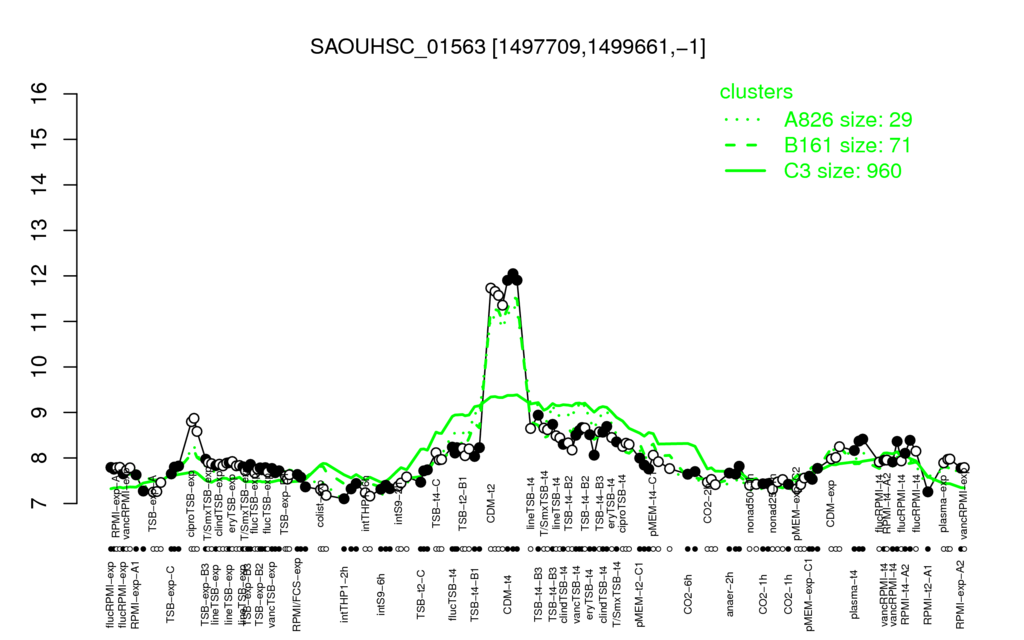 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
