Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL0455 [new locus tag: SACOL_RS02305 ]
- pan locus tag?: SAUPAN001990000
- symbol: SACOL0455
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL0455 [new locus tag: SACOL_RS02305 ]
- symbol: SACOL0455
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 457594..458550
- length: 957
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3236483 NCBI
- RefSeq: YP_185345 NCBI
- BioCyc: see SACOL_RS02305
- MicrobesOnline: 911925 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901ATGTTAACTAAAGAATTTGCACAACGCGTAGAACTAAGTGAGAAGCAAGTACGTAAAATT
GTTCAACATTTAGAAGAACGTGGTTACCAACTAAGCAAGACTGAATATCGTGGTCGTGAA
GCAACTGATTTCAAAGAAGAAGATATCGAGCTTTTCAAAGACATTGCCGACAAAGTAAAA
CAAACAAATAGTTATGATCTAGCGTTTGATGAATTAGAAAAAGAAAAAGACTTCCTGCAA
GTCATTGTCAAAAACGATGACAAAAACTTACCTACTAATCAAAATGTCGCTCAACTAGTA
GAAGATTTACGCCTAGAAATCCAGAAAATGCGCGAAGAACGTCACCTACTTGGTCAAATG
ATGAATCAAGTACATCAGCAACAACAAGAATTAAAAGAACTTCAAAATCAACTTACATCT
AAAATCGATTCAAATAGCGAATCCTTAAAAGCCATCCAAACATCACAAGAGGCTATCCAA
GAAGCGCAAGCCTCTCAAGCAAAAGCATTAGCTGAATCAACAAATAAAGTTGAAAAGAAT
GCTGTTACTGAAGATAAAGCTGACAGTAAAGATTCGAAGGTTGCTGGAGTGAATACATCA
ACAGACGCTAAGACGGATACTAAAGCAGAAAATGCAGGCGATGGCACTGCTACTAAAGTA
GATAAAGAGGATCAAATTTCTGCAACTGAAGCAATTGAAAAAGCCAGCGTTGAGCAGTCT
AAAAATGACAATGCAGCTGAGACTTCAAATAAAGAAGCTACAGTAGATGCAGAAGCGCAA
CATGATGCTGAACAACAAGTAGCAGAAGCCCATGCAGAAGCTTCTAAACAAGCTACTTCG
AACGATTCATTGGAAGCGAAAGCAGAAAATGACAGCACAGCTTCACAGTCAGAAATGTCA
GAACCAAAACCTCAAGAAGAGAAAAAAGGTTTCTTCGCACGTTTATTCAACCTGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
957
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL0455 [new locus tag: SACOL_RS02305 ]
- symbol: SACOL0455
- description: hypothetical protein
- length: 318
- theoretical pI: 4.46066
- theoretical MW: 35611.7
- GRAVY: -1.05126
⊟Function[edit | edit source]
- TIGRFAM: DNA metabolism DNA replication, recombination, and repair DNA excision repair protein (rad2) (TIGR00600; HMM-score: 3.8)Cellular processes Cell division chromosome segregation protein SMC (TIGR02168; HMM-score: 3.5)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02168; HMM-score: 3.5)Transport and binding proteins Cations and iron carrying compounds K+-dependent Na+/Ca+ exchanger (TIGR00927; HMM-score: 3.3)
- TheSEED :
- FIG01108085: hypothetical protein
- PFAM: no clan defined Med9; RNA polymerase II transcription mediator complex subunit 9 (PF07544; HMM-score: 18.5)and 4 moreHTH (CL0123) HTH_36; Helix-turn-helix domain (PF13730; HMM-score: 14.3)NirdL-like_HTH; Siroheme decarboxylase NirDL-like HTH domain (PF22451; HMM-score: 12.2)FAD-oxidase_C (CL0277) FAD_SOX; Flavin adenine dinucleotide (FAD)-dependent sulfhydryl oxidase (PF18371; HMM-score: 10.5)HTH (CL0123) HTH_54; ParA helix turn helix domain (PF18607; HMM-score: 9.4)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.8369
- Cytoplasmic Membrane Score: 0.0327
- Cell wall & surface Score: 0.1159
- Extracellular Score: 0.0145
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003324
- TAT(Tat/SPI): 0.000347
- LIPO(Sec/SPII): 0.000788
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLTKEFAQRVELSEKQVRKIVQHLEERGYQLSKTEYRGREATDFKEEDIELFKDIADKVKQTNSYDLAFDELEKEKDFLQVIVKNDDKNLPTNQNVAQLVEDLRLEIQKMREERHLLGQMMNQVHQQQQELKELQNQLTSKIDSNSESLKAIQTSQEAIQEAQASQAKALAESTNKVEKNAVTEDKADSKDSKVAGVNTSTDAKTDTKAENAGDGTATKVDKEDQISATEAIEKASVEQSKNDNAAETSNKEATVDAEAQHDAEQQVAEAHAEASKQATSNDSLEAKAENDSTASQSEMSEPKPQEEKKGFFARLFNL
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
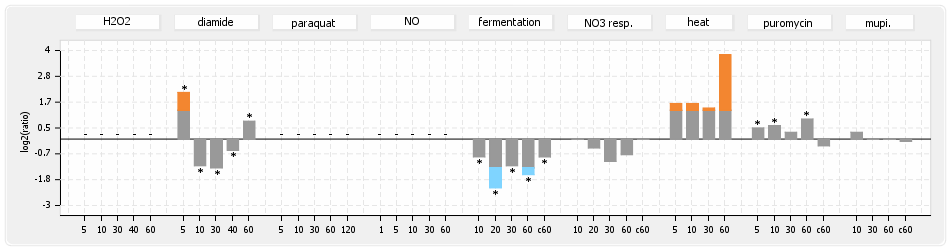
- S.aureus Expression Data Browser: data available for NCTC8325
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: 10.83 h [6]
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Jan Pané-Farré, Andreas Otto, Susanne Sievers, Michael Hecker, Dörte Becher
Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach.
J Proteome Res: 2010, 9(3);1579-90
[PubMed:20108986] [WorldCat.org] [DOI] (I p) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ Stephan Michalik, Jörg Bernhardt, Andreas Otto, Martin Moche, Dörte Becher, Hanna Meyer, Michael Lalk, Claudia Schurmann, Rabea Schlüter, Holger Kock, Ulf Gerth, Michael Hecker
Life and death of proteins: a case study of glucose-starved Staphylococcus aureus.
Mol Cell Proteomics: 2012, 11(9);558-70
[PubMed:22556279] [WorldCat.org] [DOI] (I p)