Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01583
- pan locus tag?: SAUPAN003962000
- symbol: SAOUHSC_01583
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01583
- symbol: SAOUHSC_01583
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1508540..1510567
- length: 2028
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919999 NCBI
- RefSeq: YP_500098 NCBI
- BioCyc: G1I0R-1471 BioCyc
- MicrobesOnline: 1290012 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981ATGAATAAAATTAATGATAGGGATTTAACAGAATTAAGTAGTTACTGGGTTTATCAAAAT
ATTGATATAAAAAAAGAATTTAAAGTTAATGGAAAAAGGTTTAAACAAGTAGACAGTTAT
AATGATGATAAGAATAGTAATTTGAATGGTGCTGCTGATATTAAAATATATGAGTTATTA
GATGATAAAAGTAAACCAACTGGTCAACAGACAATAATTTATCAAGGAACATCTAATGAG
GCAATTAATCCAAATAATCCATTAAAATCATCGGGGTTTGGAGATGATTGGCTCCAAAAT
GCTAAATTAATGAATAATGATAATGAAAGCACAGATTATTTAAAGCAAACAGATCAATTA
TCAAATCAATATAAAATAAAGTTAGAAGATGCAGATAGATTATCAAATAGTGATTTTTTA
AAAAAATATAGAATGGAATCAAGTAACTTCAAAAACAAAACCATTGTGGCGGATGGCGGT
AATTCGGAAGGCGGTGCAGGAGCAAAATATCAAGGAGCGAAACATCCGAATGAAAAAGTT
GTTGCTACTGACTCAGCAATGATTCCTTATGCTGCTTGGCAGAAATTTGCTAGACCACGC
TTTGATAATATGATTAGTTTTAATAGTACCAACGATTTATTAACATGGTTACAAGATCCA
TTCATCAAAGATATGCCAGGAAAACGCGTTAACATTAATGATGGTGTGCCCAGGTTAGAT
ACTTTAATAGACAGCCATGTAGGTTATAAAAGGAAGTTAAATAGAAAAGATAACACATAC
GATACTGTACCACTAATCAAAATAAAGTCGGTAAAAGATACAGAAATTAAAAATGGAAAA
AAAGTAAAAAAGACTATTAACATAACATTAGATATGGATGGGCGAATTCCAATAAATGTT
TGGACAGGAGATTCGATTGCACGTTCTGGAAGAGGAACTTTAATTAAACTTAATTTAGAA
AATCTTGATGCGTTGAGTAAACTGATTACTGGTGAAACAAGTGGTATGTTAGCAGAATGC
GTAATCTTTTTAAATGAAAGTTTTAACATCTCAGAAAATGAAAATAAAAATTTTGCAGAT
AGAAAGAAACAATTATCAGAAGGATTTAAGGATAAGATTAACTTATTTCAGTTAGAAGAA
ATGGAAAGAACTTTAATTAGTAAAATAAACTCACTTGAAGAAGTTGCAGATGAAACAATA
GAAAGTATTAGTGCTGTTAAACACTTATTACCTGATTTTGCATTGGATGCATTAAAAGAA
AGAATTAATGAGTTGTTTAAAGGTATAAAATCTTTTATAGAAAAAGTGTATGATAGTATA
GATAATGAAATTTTAGAAATTTTCAAAAATATAGATCACGACTTCAGAGATGGAGTATCT
GAAGAAATGATGAAACATTTGAAAGTAGTGAAACAGAATATAGACCAAATAAAAAATCAA
AATGATATTTATGGTAGGCAAATTGCAGATATTAGAAGTATTATGAAACAACAAGATGCA
ACAATTTTAGATGGAAATTTTCAAATTAATTGTAGCGGCGAAAATATGGTACAGGGTCTA
GTTATACCTTCTAATTATTTAGGAAGAAAAATGAAAATATTAAAAGACCATATCGATGAT
GGTATTAAAAAAATAGCAGACTATGTTCAAGGTATATATGATGAATATGCATCGAAAATT
GTCGATGTAATAAAATATTTGATTAATACAATTCCCAAAATACGTAAGAATTTAAGACAT
GCAATTGAAATGTTAAATGTAAAAAAGAAAGAATTTTTGTCCCTGATTCCTAATGTAACT
TGTAATTATATTAAAACTAAATTAGAAGAATTAGATAATACTTTAGGCAAATGGGAGCCT
TTTCTTAATGATTTAAAAGCAGTGTCACCAATTTTAGATAACCATTTAGATGATATTGTT
AAGAACATGAAGCCTTTGATTGTACAAATGATATTTGAACCATCACATTATGACGATATG
TTTAATTCAAGAAAAGCTTTAACGCCAGTGTTCTCAAGCGTTTTATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2028
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01583
- symbol: SAOUHSC_01583
- description: hypothetical protein
- length: 675
- theoretical pI: 6.81306
- theoretical MW: 77180.5
- GRAVY: -0.56637
⊟Function[edit | edit source]
- ⊞TIGRFAM: Cellular processes Detoxification formyl-CoA transferase (TIGR03253; EC 2.8.3.16; HMM-score: 16.5)and 1 more
- TheSEED :
- hypothetical protein within a prophage
- ⊞PFAM: no clan defined Baculo_PEP_C; Baculovirus polyhedron envelope protein, PEP, C terminus (PF04513; HMM-score: 24.9)and 7 more
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNKINDRDLTELSSYWVYQNIDIKKEFKVNGKRFKQVDSYNDDKNSNLNGAADIKIYELLDDKSKPTGQQTIIYQGTSNEAINPNNPLKSSGFGDDWLQNAKLMNNDNESTDYLKQTDQLSNQYKIKLEDADRLSNSDFLKKYRMESSNFKNKTIVADGGNSEGGAGAKYQGAKHPNEKVVATDSAMIPYAAWQKFARPRFDNMISFNSTNDLLTWLQDPFIKDMPGKRVNINDGVPRLDTLIDSHVGYKRKLNRKDNTYDTVPLIKIKSVKDTEIKNGKKVKKTINITLDMDGRIPINVWTGDSIARSGRGTLIKLNLENLDALSKLITGETSGMLAECVIFLNESFNISENENKNFADRKKQLSEGFKDKINLFQLEEMERTLISKINSLEEVADETIESISAVKHLLPDFALDALKERINELFKGIKSFIEKVYDSIDNEILEIFKNIDHDFRDGVSEEMMKHLKVVKQNIDQIKNQNDIYGRQIADIRSIMKQQDATILDGNFQINCSGENMVQGLVIPSNYLGRKMKILKDHIDDGIKKIADYVQGIYDEYASKIVDVIKYLINTIPKIRKNLRHAIEMLNVKKKEFLSLIPNVTCNYIKTKLEELDNTLGKWEPFLNDLKAVSPILDNHLDDIVKNMKPLIVQMIFEPSHYDDMFNSRKALTPVFSSVL
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [1] : S636 < SAOUHSC_01583 < SAOUHSC_01584 < S637
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
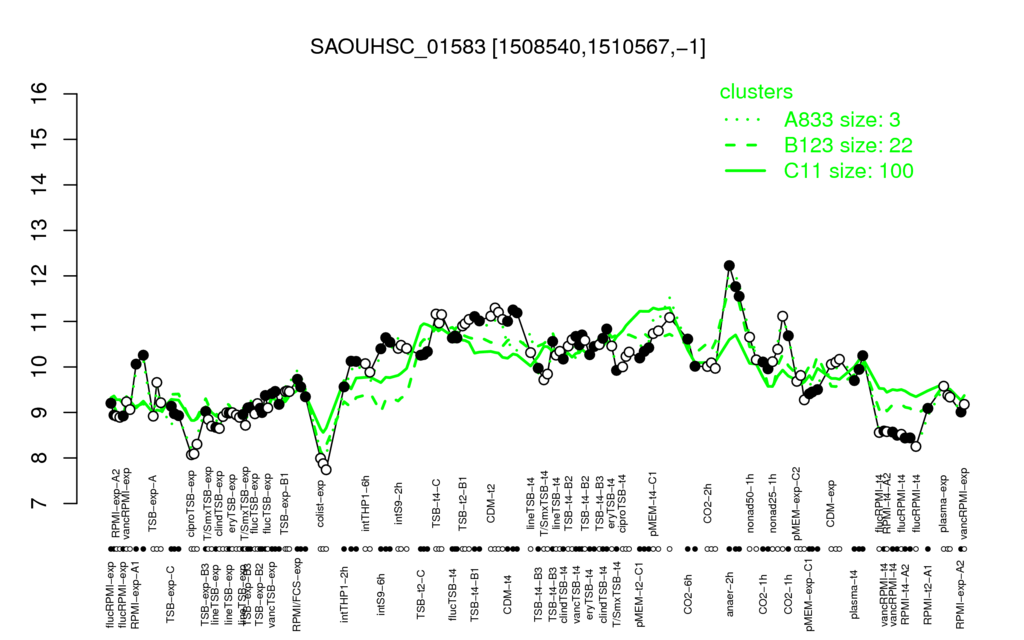 ⊟Multi-gene expression profiles
⊟Multi-gene expression profiles
Click on any data point to display a description of the corresponding condition!
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊞Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊞Other Information[edit | edit source]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Jump up to: 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
