Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02297
- pan locus tag?: SAUPAN005331000
- symbol: SAOUHSC_02297
- pan gene symbol?: —
- synonym:
- product: S1 RNA-binding domain-containing protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02297
- symbol: SAOUHSC_02297
- product: S1 RNA-binding domain-containing protein
- replicon: chromosome
- strand: -
- coordinates: 2129345..2131495
- length: 2151
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919168 NCBI
- RefSeq: YP_500775 NCBI
- BioCyc: G1I0R-2169 BioCyc
- MicrobesOnline: 1290736 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101ATGGACAATCAATTGATTAATTCAATCATAGAGAAATATCAATTTAGTAAAAAACAAATT
GAAGCAGTATTAACACTGCTAGAAGAAAAAAATACAGTACCATTTATTGCGAGGTATCGA
AAAGAGCAAACTGGTGGACTAGATGAAGTTCAAATAAAGCAAATTGATGACGAATACCAA
TATATGGTCAATTTACAAAAACGTAAAGAAGAAGTTATCAAAAATATAGAACAGCAAGGA
TTACTTACTGAGGAATTAAAGAAGGATATTTTAAAACAGAACAAATTACAACGTGTTGAA
GACCTATATAGGCCTTTTAAACAAAAGAAAAAGACAAGGGCAACTGAGGCGAAACGTAAA
GGGTTAGAGCCATTAGCGATATGGATGAAGGCACGTAAACATGAAGTCTCAATTGAAGAA
AAAGCACAACAATTTATAAATGAAGAAGTGCAATCGGTTGAAGATGCTATCAAAGGTGCA
CAAGATATTATTGCGGAACAAATTTCAGATAATCCTAAATATAGAACAAAAATTTTAAAA
GATATGTATCATCAAGGTGTGTTAACTACATCTAAAAAGAAAAATGCTGAAGATGAAAAA
GGTATTTTTGAAATGTACTATGCATATAGTGAGCCAATTAAACGCATTGCTAATCATAGA
GTTTTAGCTGTTAATCGTGGTGAAAAAGAGAAAGTATTATCTGTAAAGTTTGAATTCGAT
ACGACATCAGTAGAGGATTTCATTGCACGTCAAGAAATCAATCATAATAATGTAAATCGC
AGTTATATTTTAGAGGCGATTAAAGATAGCTTGAAACGCTTAATTGTCCCTTCGATAGAG
CGTGAAATCCATGCTGATTTAACTGAAAAAGCTGAAAATCATGCAATAGATGTTTTTAGT
GAAAACTTAAGAAATCTATTACTGCAACCTCCAATGAAAGGTAAACAAATATTAGGCGTA
GATCCAGCATTTAGAACAGGTTGTAAATTAGCAGTCATTAACCCATTCGGTACTTTTATA
GCAAAAGGTGTGATTTATCCGCATCCACCAGTTTCTAAAAAAGAGGCAGCAGAGAAGGAT
TTTGTACAAATGGTTAAAGCGTATGATGTGCAATTAATTGCAATTGGCAATGGTACTGCA
AGTCGTGAAACAGAACAATTTGTTGCAGATTTAATTAAAAAGCATCAGTTGCCAGTACAA
TTCATCATTGTCAATGAAGCGGGCGCTTCAGTATACTCAGCATCAGAAATTGCTAGAGAT
GAATTTCCTGATTTTCAAGTGGAAGAAAGAAGTGCCGTATCAATTGGACGACGCGTTCAA
GATCCATTAAGTGAATTAGTTAAAATCGATCCTAAATCTATTGGTGTCGGACAATACCAG
CATGATGTTAATCAAAAAGCACTAGAAAATGCATTAACTTTCGTTGTTGAAACAGCAGTT
AACCAAGTAGGTGTTGATGTCAATACAGCATCATCATCATTATTACAATATGTGTCTGGT
CTAAGTTCGCAAATTGCTAAAAATATAATTGCATATAGAGAAGAAAATGGCGCAATAAAG
CATAATAAAGAGTTAAGCAAGATTAAGCGTTTGGGTGCTAAAACTTTTGAGCAGAGTATA
GGATTTTTAAGAATAGTAGACGGTTCAGAGCCATTGGATAATACATCGATTCATCCCGAA
AGCTATAAAGTAACGTATCAGTTATTAGATAAACTCGGATTTGGTGGCAATGATTTAGGT
AGTGATGCATTAAAAGCTAAATTAAATTCATTGGATATGGATGAACTGGCGATTGAATTA
CAAGTCGGTGTACCTACATTAGAGGATATTATTAAATCCTTGAAAGCACCTAATCGAGAT
CCAAGGGACGAGTTTGAAACACCAATCCTTAAATCCGATGTGCTATCAATTGAAGATTTA
CAAGAAGGAATGAAATTGAGTGGTACAGTTAGAAATGTAGTAGATTTTGGAGCATTTGTA
GATATAGGTGTTAAACAAGATGGACTTGTCCATGTTTCGAAACTTTCTAAGAAGTTCGTA
AAAAATCCAATGGATATTGTCAGTGTTGGTGATATCGTTGACGTTTGGATTTACAGTATT
GACAAAAATAAAGACAAAGTATCATTAACGATGATTGATCCACATGAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2151
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02297
- symbol: SAOUHSC_02297
- description: S1 RNA-binding domain-containing protein
- length: 716
- theoretical pI: 6.24525
- theoretical MW: 80933.9
- GRAVY: -0.426397
⊟Function[edit | edit source]
- TIGRFAM: Protein synthesis Ribosomal proteins: synthesis and modification ribosomal protein bS1 (TIGR00717; HMM-score: 55.9)Transcription Degradation of RNA polyribonucleotide nucleotidyltransferase (TIGR03591; EC 2.7.7.8; HMM-score: 49)and 5 moreCellular processes DNA transformation comEA protein (TIGR01259; HMM-score: 40.9)Cellular processes DNA transformation competence protein ComEA helix-hairpin-helix repeat region (TIGR00426; HMM-score: 36.6)guanosine pentaphosphate synthetase I/polyribonucleotide nucleotidyltransferase (TIGR02696; EC 2.7.-.-,2.7.7.8; HMM-score: 26)Mobile and extrachromosomal element functions Prophage functions phage uncharacterized protein, C-terminal domain (TIGR01630; HMM-score: 16.1)Transcription DNA-dependent RNA polymerase DNA-directed RNA polymerase (TIGR00448; EC 2.7.7.6; HMM-score: 15.4)
- TheSEED :
- Transcription accessory protein (S1 RNA-binding domain)
and 1 more - PFAM: YqgF (CL0580) Tex_YqgF; Tex protein YqgF-like domain (PF16921; HMM-score: 182.4)and 11 moreno clan defined Tex_central_region; Tex central region-like (PF22706; HMM-score: 102.2)PWI-like (CL0805) Tex_N; Tex-like protein N-terminal domain (PF09371; HMM-score: 101.1)HHH (CL0198) HHH_3; Helix-hairpin-helix motif (PF12836; HMM-score: 92.7)HHH_9; HHH domain (PF17674; HMM-score: 75.7)OB (CL0021) S1; S1 RNA binding domain (PF00575; HMM-score: 56.8)YqgF (CL0580) YqgF; Holliday-junction resolvase-like of SPT6 (PF14639; HMM-score: 33.3)HHH (CL0198) HHH_7; Helix-hairpin-helix motif (PF14635; HMM-score: 32.2)OB (CL0021) S1_RRP5; RRP5 S1 domain (PF23459; HMM-score: 20.9)HHH (CL0198) T2SSK; Type II secretion system (T2SS), protein K (PF03934; HMM-score: 16.7)no clan defined T4_RIIA1; RIIA.1-like protein (PF24210; HMM-score: 13.9)SRA1; Steroid receptor RNA activator (SRA1) (PF07304; HMM-score: 12.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.973
- Cytoplasmic Membrane Score: 0.0129
- Cell wall & surface Score: 0.0063
- Extracellular Score: 0.0078
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.001492
- TAT(Tat/SPI): 0.000112
- LIPO(Sec/SPII): 0.000284
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MDNQLINSIIEKYQFSKKQIEAVLTLLEEKNTVPFIARYRKEQTGGLDEVQIKQIDDEYQYMVNLQKRKEEVIKNIEQQGLLTEELKKDILKQNKLQRVEDLYRPFKQKKKTRATEAKRKGLEPLAIWMKARKHEVSIEEKAQQFINEEVQSVEDAIKGAQDIIAEQISDNPKYRTKILKDMYHQGVLTTSKKKNAEDEKGIFEMYYAYSEPIKRIANHRVLAVNRGEKEKVLSVKFEFDTTSVEDFIARQEINHNNVNRSYILEAIKDSLKRLIVPSIEREIHADLTEKAENHAIDVFSENLRNLLLQPPMKGKQILGVDPAFRTGCKLAVINPFGTFIAKGVIYPHPPVSKKEAAEKDFVQMVKAYDVQLIAIGNGTASRETEQFVADLIKKHQLPVQFIIVNEAGASVYSASEIARDEFPDFQVEERSAVSIGRRVQDPLSELVKIDPKSIGVGQYQHDVNQKALENALTFVVETAVNQVGVDVNTASSSLLQYVSGLSSQIAKNIIAYREENGAIKHNKELSKIKRLGAKTFEQSIGFLRIVDGSEPLDNTSIHPESYKVTYQLLDKLGFGGNDLGSDALKAKLNSLDMDELAIELQVGVPTLEDIIKSLKAPNRDPRDEFETPILKSDVLSIEDLQEGMKLSGTVRNVVDFGAFVDIGVKQDGLVHVSKLSKKFVKNPMDIVSVGDIVDVWIYSIDKNKDKVSLTMIDPHE
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [3] (data from MRSA252) SAOUHSC_02477 (rpsI) 30S ribosomal protein S9 [3] (data from MRSA252) SAOUHSC_01040 pyruvate dehydrogenase complex, E1 component subunit alpha [3] (data from MRSA252) SAOUHSC_02969 arginine deiminase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_02296 < SAOUHSC_02297predicted SigA promoter [4] : S888 < SAOUHSC_02296 < SAOUHSC_02297 < S889 < SAOUHSC_02298 < SAOUHSC_02299 < SAOUHSC_02300 < S890 < SAOUHSC_02301 < S891 < SAOUHSC_02303 < SAOUHSC_02304
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
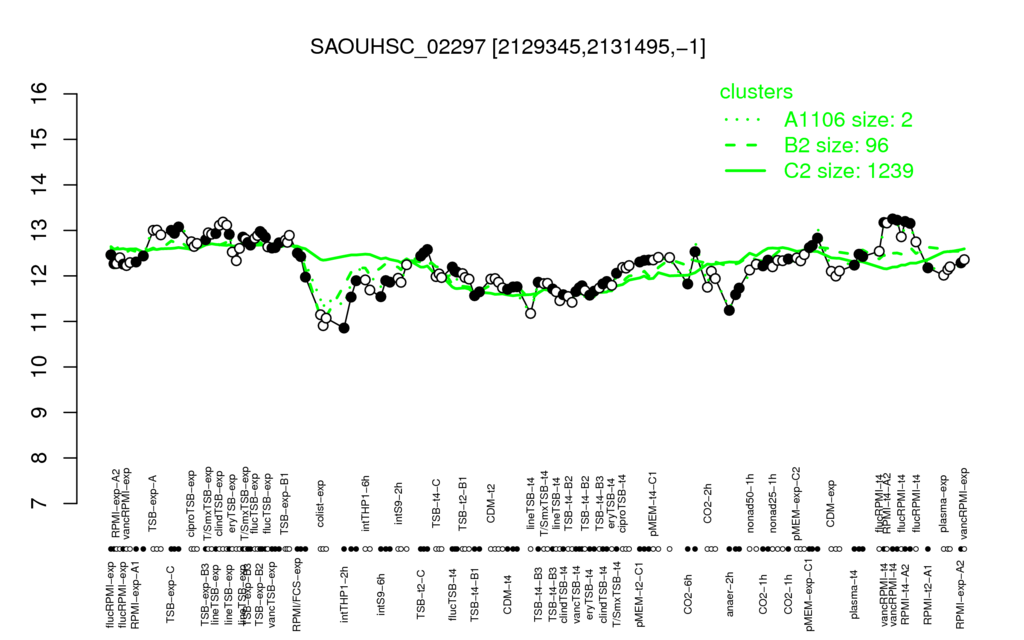 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 3.3 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
