Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02009
- pan locus tag?: SAUPAN004854000
- symbol: SAOUHSC_02009
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02009
- symbol: SAOUHSC_02009
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1918269..1919786
- length: 1518
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921889 NCBI
- RefSeq: YP_500506 NCBI
- BioCyc: G1I0R-1903 BioCyc
- MicrobesOnline: 1290462 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501ATGGGAAGTTCAATTGTTTTAAAGTTACTAAAAGTAACACACTACTATAGGAATAAACAG
AATAAGAAATGGTATTTACCTTTTGGATATGATGCTGAAGATATCGATTTAAACAATATT
AGTTTACATATTTATCAAGGAGAAGCATTAGGTATAATTGGTGAACCTGAATCTTCCAAA
GCGTTGGTAGGTCAATTGTTGGCAGGTGCAATTAAACCTGATAAAGGTAAAGTAGTTTGT
ACTGAAGATTTGTTCTACGGATATATTGAAGATCAATCGTTAATTCATCAAACTGTTGAA
GCTTATACAGCGCAGTTAGTTCAACTATTTCCATATGAAATTAATGATCATAAAGCTGAA
CAGATTATTCAATATGCACATTTAGGTGATTATAAAACGAAGCCGGTTAACCATATTTCG
AAAGCGGCATACGCTCAATTACTATTAAGTATTGCACGCTCATCAAAATCAAATATTATT
ATTTTAAATCATGTTATTGACTATTTAACACCACAATTTATGGAACGTGCGATTGAATTA
ACAAATGATTATATTGAAAATAATTTAACGATTGTGTCAATTGGTGATGATATTGATAAA
ATTTCACAAGTGAGTAACTACATAGCTTGGTTTTCACATGGTCAATTAAGAATGGAAGGG
TCACTTAAACAAGTTATTCCATCTTTTAAAGAACATGAACGTGATCGATTAAGTCTAAAC
TCAAAAGAAGAAATTGAAAACTTTGATTTAGATTGGAAAAAGAATCGTACAAGAATACCA
GAGATGACCTATAATTTCAAACGTGTTGAGCGCTATAATCATGCAAAACCGCCTAAGTTT
TTAGTGCGTTTTTGGACTTTAGCCTCAGGTACTATTTTAGGCTTAGCATTGATGATGTTG
CTCATTTTCAATAATATAGGAATTATTTCGATAACAGATTTTACGAATCGTGCTACGATG
CAAAATGAAAATAAAGATCCATATGGCGAAAAGTTAGCTTATGGAATTGCTTTTAATGGC
AGTGTGGATATGCAAGGGGATAAACAAGTCACAATTCCAAAATATAGTGTAGTTACAATT
ACTGGCGAAAATAGTAAAAATTATCGTGTTACCGCCGATAATAAGACTTACTATGTTAGT
AAAGATAAATTAGAATATTTTAACCCGGCAGGTTTATATCAAACGCATAGTTTTAAAAAA
TTAGCACCATATATGAAATCAAATTATAGTAATTACTATGCATACTTTAATAGTCAATTA
CATAAAAAGCATAGTTCAGTTATAAAAACTTTAGTTCCTGATGATGATAACCGTTTCGTT
GCATCCGTTACACAACAACCGATACAATTACTTTTCAATGATAATAATCAGTTATACGGT
TTTGTTTATCCAATTGTAGATAAAAAAGAATTAAAAGATAAGTTTAATATTAACAATAAC
ATTTGGATTACTAAAGTTGGGAATGGATATTGTATTGCCAATTTGAAAGAAGACAAATGG
ATTTATATTGAATTGTAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1518
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02009
- symbol: SAOUHSC_02009
- description: hypothetical protein
- length: 505
- theoretical pI: 8.41277
- theoretical MW: 58254
- GRAVY: -0.39604
⊟Function[edit | edit source]
- TIGRFAM: Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04521; EC 3.6.3.-; HMM-score: 43.2)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04520; EC 3.6.3.-; HMM-score: 41)and 43 morelantibiotic protection ABC transporter, ATP-binding subunit (TIGR03740; HMM-score: 32.9)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtD (TIGR03411; HMM-score: 30.7)Transport and binding proteins Other daunorubicin resistance ABC transporter, ATP-binding protein (TIGR01188; HMM-score: 29.1)Energy metabolism Methanogenesis methyl coenzyme M reductase system, component A2 (TIGR03269; HMM-score: 26)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikE (TIGR02769; EC 3.6.3.24; HMM-score: 25.5)Cellular processes Pathogenesis type I secretion system ATPase (TIGR03375; HMM-score: 24.1)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR03375; HMM-score: 24.1)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 23.5)Transport and binding proteins Other lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 23.5)Transport and binding proteins Cations and iron carrying compounds cobalt ABC transporter, ATP-binding protein (TIGR01166; HMM-score: 23.2)Transport and binding proteins Amino acids, peptides and amines glycine betaine/L-proline transport ATP binding subunit (TIGR01186; HMM-score: 22.8)Transport and binding proteins Anions phosphate ABC transporter, ATP-binding protein (TIGR00972; EC 3.6.3.27; HMM-score: 22.7)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01842; HMM-score: 22.3)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01846; HMM-score: 20.7)Protein fate Protein and peptide secretion and trafficking lipoprotein releasing system, ATP-binding protein (TIGR02211; EC 3.6.3.-; HMM-score: 20.3)2-aminoethylphosphonate ABC transport system, ATP-binding component PhnT (TIGR03258; HMM-score: 20.2)D-methionine ABC transporter, ATP-binding protein (TIGR02314; EC 3.6.3.-; HMM-score: 19)Transport and binding proteins Anions nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 18.9)Transport and binding proteins Other nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 18.9)ATP-binding cassette protein, ChvD family (TIGR03719; HMM-score: 18.1)Cellular processes Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 17.7)Transport and binding proteins Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 17.7)thiol reductant ABC exporter, CydC subunit (TIGR02868; HMM-score: 17.3)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 17.2)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 17.2)Transport and binding proteins Other thiamine ABC transporter, ATP-binding protein (TIGR01277; EC 3.6.3.-; HMM-score: 16.6)ectoine/hydroxyectoine ABC transporter, ATP-binding protein EhuA (TIGR03005; HMM-score: 16.1)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 15.5)Transport and binding proteins Other LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 15.5)Transport and binding proteins Carbohydrates, organic alcohols, and acids D-xylose ABC transporter, ATP-binding protein (TIGR02633; EC 3.6.3.17; HMM-score: 15.2)Transport and binding proteins Anions sulfate ABC transporter, ATP-binding protein (TIGR00968; EC 3.6.3.25; HMM-score: 14.6)Central intermediary metabolism Phosphorus compounds phosphonate C-P lyase system protein PhnK (TIGR02323; HMM-score: 14.5)Transport and binding proteins Anions phosphonate ABC transporter, ATP-binding protein (TIGR02315; EC 3.6.3.28; HMM-score: 14.3)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 14.2)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 14.2)Protein fate Protein and peptide secretion and trafficking heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 13.6)Transport and binding proteins Other heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 13.6)gliding motility-associated ABC transporter ATP-binding subunit GldA (TIGR03522; HMM-score: 13.2)Cellular processes Toxin production and resistance putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 13)Transport and binding proteins Unknown substrate putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 13)thiol reductant ABC exporter, CydD subunit (TIGR02857; HMM-score: 12.3)Cellular processes Cell division cell division ATP-binding protein FtsE (TIGR02673; HMM-score: 12.2)proposed F420-0 ABC transporter, ATP-binding protein (TIGR03873; HMM-score: 11.1)
- TheSEED :
- Teichoic acid export ATP-binding protein TagH (EC 7.5.2.4)
- PFAM: no clan defined TagH_C; Teichoic acid transporter subunit TagH C-terminal domain (PF22096; HMM-score: 141.6)and 2 moreSH3 (CL0010) TagH_SH3-like; Teichoic acid transporter subunit TagH, SH3-like domain (PF22269; HMM-score: 108.2)P-loop_NTPase (CL0023) ABC_tran; ABC transporter (PF00005; HMM-score: 32)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: unknown (no significant prediction)
- Cytoplasmic Score: 2.5
- Cytoplasmic Membrane Score: 2.5
- Cellwall Score: 2.5
- Extracellular Score: 2.5
- Internal Helix: 1
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0005
- Cytoplasmic Membrane Score: 0.9913
- Cell wall & surface Score: 0.0003
- Extracellular Score: 0.0079
- LocateP: Intracellular /TMH start AFTER 60
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: Possibly Sec-
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.005976
- TAT(Tat/SPI): 0.000099
- LIPO(Sec/SPII): 0.00046
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MGSSIVLKLLKVTHYYRNKQNKKWYLPFGYDAEDIDLNNISLHIYQGEALGIIGEPESSKALVGQLLAGAIKPDKGKVVCTEDLFYGYIEDQSLIHQTVEAYTAQLVQLFPYEINDHKAEQIIQYAHLGDYKTKPVNHISKAAYAQLLLSIARSSKSNIIILNHVIDYLTPQFMERAIELTNDYIENNLTIVSIGDDIDKISQVSNYIAWFSHGQLRMEGSLKQVIPSFKEHERDRLSLNSKEEIENFDLDWKKNRTRIPEMTYNFKRVERYNHAKPPKFLVRFWTLASGTILGLALMMLLIFNNIGIISITDFTNRATMQNENKDPYGEKLAYGIAFNGSVDMQGDKQVTIPKYSVVTITGENSKNYRVTADNKTYYVSKDKLEYFNPAGLYQTHSFKKLAPYMKSNYSNYYAYFNSQLHKKHSSVIKTLVPDDDNRFVASVTQQPIQLLFNDNNQLYGFVYPIVDKKELKDKFNINNNIWITKVGNGYCIANLKEDKWIYIEL
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
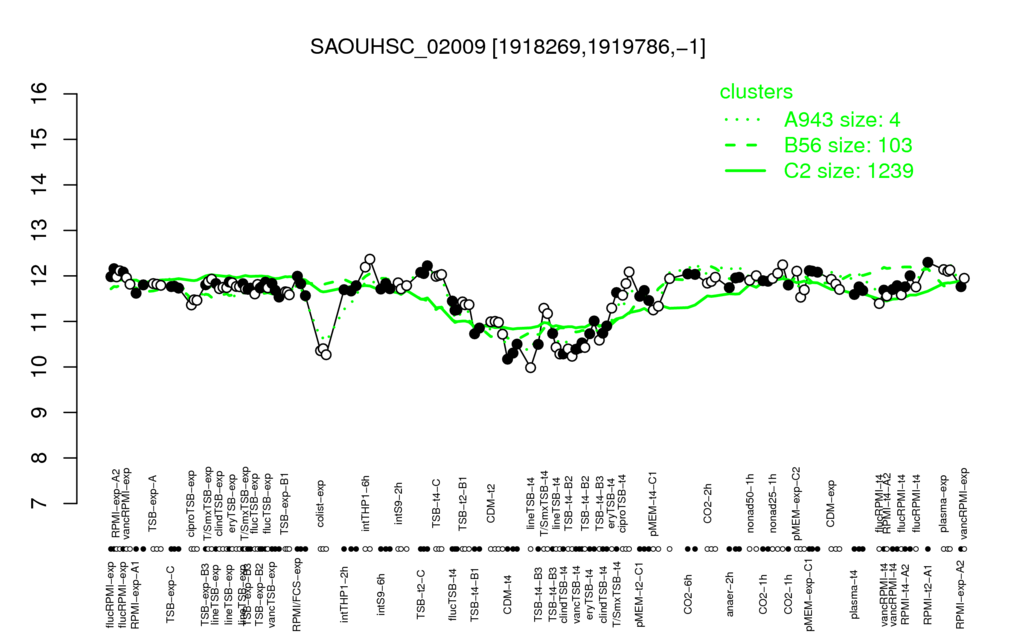 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
