Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01843
- pan locus tag?: SAUPAN004393000
- symbol: SAOUHSC_01843
- pan gene symbol?: isdH
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01843
- symbol: SAOUHSC_01843
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1747419..1750106
- length: 2688
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920522 NCBI
- RefSeq: YP_500348 NCBI
- BioCyc: G1I0R-1714 BioCyc
- MicrobesOnline: 1290262 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641ATGAACAAACATCACCCAAAATTAAGGTCTTTCTATTCTATTAGAAAATCAACTCTAGGC
GTTGCATCGGTCATTGTCAGTACACTATTTTTAATTACTTCTCAACATCAAGCACAAGCA
GCAGAAAATACAAATACTTCAGATAAAATCTCGGAAAATCAAAATAATAATGCAACTACA
ACTCAGCCACCTAAGGATACAAATCAAACACAACCTGCTACGCAACCAGCAAACACTGCG
AAAAACTATCCTGCAGCGGATGAATCACTTAAAGATGCAATTAAAGATCCTGCATTAGAA
AATAAAGAACATGATATAGGTCCAAGAGAACAAGTCAATTTCCAGTTATTAGATAAAAAC
AATGAAACGCAGTACTATCACTTTTTCAGCATCAAAGATCCAGCAGATGTGTATTACACT
AAAAAGAAAGCAGAAGTTGAATTAGACATCAATACTGCTTCAACATGGAAGAAGTTTGAA
GTCTATGAAAACAATCAAAAATTGCCAGTGAGACTTGTATCATATAGTCCTGTACCAGAA
GACCATGCCTATATTCGATTCCCAGTTTCAGATGGCACACAAGAATTGAAAATTGTTTCT
TCGACTCAAATTGATGATGGAGAAGAAACAAATTATGATTATACTAAATTAGTATTTGCT
AAACCTATTTATAACGATCCTTCACTTGTAAAATCAGATACAAATGATGCAGTAGTAACG
AATGATCAATCAAGTTCAGTCGCAAGTAATCAAACAAACACGAATACATCTAATCAAAAT
ATATCAACGATCAACAATGCTAATAATCAACCGCAGGCAACGACCAATATGAGTCAACCT
GCACAACCAAAATCGTCAACGAATGCAGATCAAGCGTCAAGCCAACCAGCTCATGAAACA
AATTCTAATGGTAATACTAACGATAAAACGAATGAGTCAAGTAATCAGTCGGATGTTAAT
CAACAGTATCCACCAGCAGATGAATCACTACAAGATGCAATTAAAAACCCGGCTATCATC
GATAAAGAACATACAGCTGATAATTGGCGACCAATTGATTTTCAAATGAAAAATGATAAA
GGTGAAAGACAGTTCTATCATTATGCTAGTACTGTTGAACCAGCAACTGTCATTTTTACA
AAAACAGGACCAATAATTGAATTAGGTTTAAAGACAGCTTCAACATGGAAGAAATTTGAA
GTTTATGAAGGTGACAAAAAGTTACCAGTCGAATTAGTATCATATGATTCTGATAAAGAT
TATGCCTATATTCGTTTCCCAGTATCTAATGGTACGAGAGAAGTTAAAATTGTGTCATCT
ATTGAATATGGTGAGAACATCCATGAAGACTATGATTATACGCTAATGGTCTTTGCACAG
CCTATTACTAATAACCCAGACGACTATGTGGATGAAGAAACATACAATTTACAAAAATTA
TTAGCTCCGTATCACAAAGCTAAAACGTTAGAAAGACAAGTTTATGAATTAGAAAAATTA
CAAGAGAAATTGCCAGAAAAATATAAGGCGGAATATAAAAAGAAATTAGATCAAACTAGA
GTAGAGTTAGCTGATCAAGTTAAATCAGCAGTGACGGAATTTGAAAATGTTACACCTACA
AATGATCAATTAACAGATTTACAAGAAGCGCATTTTGTTGTTTTTGAAAGTGAAGAAAAT
AGTGAGTCAGTTATGGACGGCTTTGTTGAACATCCATTCTATACAGCAACTTTAAATGGT
CAAAAATATGTAGTGATGAAAACAAAGGATGACAGTTACTGGAAAGATTTAATTGTAGAA
GGTAAACGTGTCACTACTGTTTCTAAAGATCCTAAAAATAATTCTAGAACGCTGATTTTC
CCATATATACCTGACAAAGCAGTTTACAATGCGATTGTTAAAGTCGTTGTGGCAAACATT
GGTTATGAAGGTCAATATCATGTCAGAATTATAAATCAGGATATCAATACAAAAGATGAT
GATACATCACAAAATAACACGAGTGAACCGCTAAATGTACAAACAGGACAAGAAGGTAAG
GTTGCTGATACAGATGTAGCTGAAAATAGCAGCACTGCAACAAATCCTAAAGATGCGTCT
GATAAAGCAGATGTGATAGAACCAGAGTCTGACGTGGTTAAAGATGCTGATAATAATATT
GATAAAGATGTGCAACATGATGTTGATCATTTATCCGATATGTCGGATAATAATCACTTC
GATAAATATGATTTAAAAGAAATGGATACTCAAATTGCCAAAGATACTGATAGAAATGTG
GATAAAGATGCCGATAATAGCGTTGGTATGTCATCTAATGTCGATACTGATAAAGACTCT
AATAAAAATAAAGACAAAGTCATACAGCTGAATCATATTGCCGATAAAAATAATCATACT
GGAAAAGCAGCAAAGCTTGACGTAGTGAAACAAAATTATAATAATACAGACAAAGTTACT
GACAAAAAAACAACTGAACATCTGCCGAGTGATATTCATAAAACTGTAGATAAAACAGTG
AAAACAAAAGAAAAAGCCGGCACACCATCGAAAGAAAACAAACTTAGTCAATCTAAAATG
CTACCAAAAACTGGAGAAACAACTTCAAGCCAATCATGGTGGGGCTTATATGCGTTATTA
GGTATGTTAGCTTTATTCATTCCTAAATTCAGAAAAGAATCTAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2688
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01843
- symbol: SAOUHSC_01843
- description: hypothetical protein
- length: 895
- theoretical pI: 4.84648
- theoretical MW: 100946
- GRAVY: -0.938101
⊟Function[edit | edit source]
- ⊞TIGRFAM: haptoglobin-binding heme uptake protein HarA (TIGR03658; HMM-score: 2117.2)and 4 more
- TheSEED :
- Cell surface receptor IsdH for hemoglobin-haptoglobin complexes
- ⊞PFAM: E-set (CL0159) NEAT; Iron Transport-associated domain (PF05031; HMM-score: 134.4)no clan defined Isd_H_B_linker; Iron-regulated surface determinant protein H/B, linker domain (PF20861; HMM-score: 129.5)and 3 more
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNKHHPKLRSFYSIRKSTLGVASVIVSTLFLITSQHQAQAAENTNTSDKISENQNNNATTTQPPKDTNQTQPATQPANTAKNYPAADESLKDAIKDPALENKEHDIGPREQVNFQLLDKNNETQYYHFFSIKDPADVYYTKKKAEVELDINTASTWKKFEVYENNQKLPVRLVSYSPVPEDHAYIRFPVSDGTQELKIVSSTQIDDGEETNYDYTKLVFAKPIYNDPSLVKSDTNDAVVTNDQSSSVASNQTNTNTSNQNISTINNANNQPQATTNMSQPAQPKSSTNADQASSQPAHETNSNGNTNDKTNESSNQSDVNQQYPPADESLQDAIKNPAIIDKEHTADNWRPIDFQMKNDKGERQFYHYASTVEPATVIFTKTGPIIELGLKTASTWKKFEVYEGDKKLPVELVSYDSDKDYAYIRFPVSNGTREVKIVSSIEYGENIHEDYDYTLMVFAQPITNNPDDYVDEETYNLQKLLAPYHKAKTLERQVYELEKLQEKLPEKYKAEYKKKLDQTRVELADQVKSAVTEFENVTPTNDQLTDLQEAHFVVFESEENSESVMDGFVEHPFYTATLNGQKYVVMKTKDDSYWKDLIVEGKRVTTVSKDPKNNSRTLIFPYIPDKAVYNAIVKVVVANIGYEGQYHVRIINQDINTKDDDTSQNNTSEPLNVQTGQEGKVADTDVAENSSTATNPKDASDKADVIEPESDVVKDADNNIDKDVQHDVDHLSDMSDNNHFDKYDLKEMDTQIAKDTDRNVDKDADNSVGMSSNVDTDKDSNKNKDKVIQLNHIADKNNHTGKAAKLDVVKQNYNNTDKVTDKKTTEHLPSDIHKTVDKTVKTKEKAGTPSKENKLSQSKMLPKTGETTSSQSWWGLYALLGMLALFIPKFRKESK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
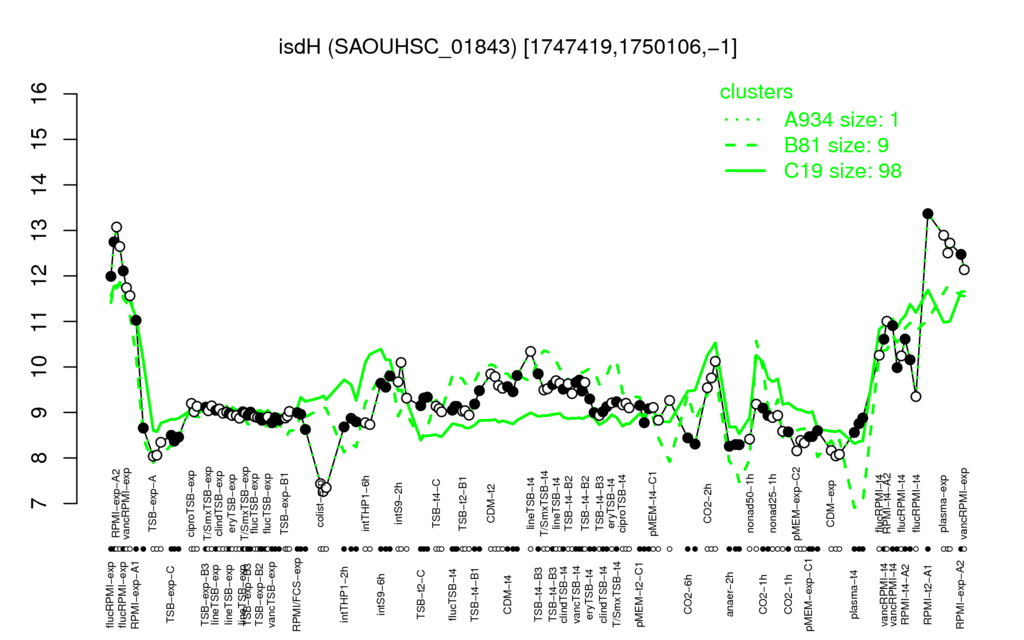 ⊟Multi-gene expression profiles
⊟Multi-gene expression profiles
Click on any data point to display a description of the corresponding condition!
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊞Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊞Other Information[edit | edit source]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Agnieszka Dryla, Dieter Gelbmann, Alexander von Gabain, Eszter Nagy
Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity.
Mol Microbiol: 2003, 49(1);37-53
[PubMed:12823809] [WorldCat.org] [DOI] (P p)
