NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02802
- pan locus tag?: SAUPAN006097000
- symbol: SAOUHSC_02802
- pan gene symbol?: fnbB
- synonym:
- product: fibronectin binding protein B
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02802
- symbol: SAOUHSC_02802
- product: fibronectin binding protein B
- replicon: chromosome
- strand: -
- coordinates: 2577879..2580632
- length: 2754
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921456 NCBI
- RefSeq: YP_501261 NCBI
- BioCyc: G1I0R-2640 BioCyc
- MicrobesOnline: 1291232 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701ATGATCGTTGTTGGAATGGGACAAGAAAAAGAAGCTGCAGCATCGGAACAAAACAATACT
ACAGTAGAGGAAAGTGGGAGTTCAGCTACTGAAAGTAAAGCAAGCGAAACACAAACAACT
ACAAATAACGTTAATACAATAGATGAAACACAATCATACAGCGCGACATCAACTGAGCAA
CCATCACAATCAACACAAGTAACAACAGAAGAAGCACCGAAAACTGTGCAAGCACCAAAA
GTAGAAACTTCGCGAGTTGATTTGCCATCGGAAAAAGTTGCTGATAAGGAAACTACAGGA
ACTCAAGTTGACATAGCTCAACCAAGTAACGTCTCAGAAATTAAACCAAGAATGAAAAGA
TCAACTGACGTTACAGCAGTTGCAGAGAAAGAAGTAGTGGAAGAAACTAAAGCGACAGGT
ACAGATGTAACAAATAAAGTGGAAGTAGAAGAAGGTAGTGAAATTGTAGGACATAAACAA
GATACGAATGTTGTAAATCCTCATAACGCAGAAAGAGTAACCTTGAAATATAAATGGAAA
TTTGGAGAAGGAATTAAGGCGGGAGATTATTTTGATTTCACATTAAGCGATAATGTTGAA
ACTCATGGTATCTCAACACTGCGTAAAGTTCCGGAGATAAAAAGTACAGATGGTCAAGTT
ATGGCGACAGGAGAAATAATTGGAGAAAGAAAAGTTAGATATACGTTTAAAGAATATGTA
CAAGAAAAGAAAGATTTAACTGCTGAATTATCTTTAAATCTATTTATTGATCCTACAACA
GTGACGCAAAAAGGTAACCAAAATGTTGAAGTTAAATTGGGTGAGACTACGGTTAGCAAA
ATATTTAATATTCAATATTTAGGTGGAGTTAGAGATAATTGGGGAGTAACAGCTAATGGT
CGAATTGATACTTTAAATAAAGTAGATGGGAAATTTAGTCATTTTGCGTACATGAAACCT
AACAACCAGTCGTTAAGCTCTGTGACAGTAACTGGTCAAGTAACTAAAGGAAATAAACCA
GGGGTTAATAATCCAACAGTTAAGGTATATAAACACATTGGTTCAGACGATTTAGCTGAA
AGCGTATATGCAAAGCTTGATGATGTCAGCAAATTTGAAGATGTGACTGATAATATGAGT
TTAGATTTTGATACTAATGGTGGTTATTCTTTAAACTTTAATAATTTAGACCAAAGTAAA
AATTATGTAATAAAATATGAAGGGTATTATGATTCAAATGCTAGCAACTTAGAATTTCAA
ACACACCTTTTTGGATATTATAACTATTATTATACAAGTAATTTAACTTGGAAAAATGGC
GTTGCATTTTACTCTAATAACGCTCAAGGCGACGGCAAAGATAAACTAAAGGAACCTATT
ATAGAACATAGTACTCCTATCGAACTTGAATTTAAATCAGAGCCGCCAGTGGAGAAGCAT
GAATTGACTGGTACAATCGAAGAAAGTAATGATTCTAAGCCAATTGATTTTGAATATCAT
ACAGCTGTTGAAGGTGCAGAAGGTCATGCAGAAGGTACCATTGAAACTGAAGAAGATTCT
ATTCATGTAGACTTTGAAGAATCGACACATGAAAATTCAAAACATCATGCTGATGTTGTT
GAATATGAAGAAGATACAAACCCAGGTGGTGGTCAGGTTACTACTGAGTCTAACCTAGTT
GAATTTGACGAAGATTCTACAAAAGGTATTGTAACTGGTGCTGTTAGCGATCATACAACA
ATTGAAGATACGAAAGAATATACGACTGAAAGTAATCTGATTGAACTAGTAGATGAACTA
CCTGAAGAACATGGTCAAGCGCAAGGACCAATCGAGGAAATTACTGAAAACAATCATCAT
ATTTCTCATTCTGGTTTAGGAACTGAAAATGGTCACGGTAATTATGGCGTGATTGAAGAA
ATCGAAGAAAATAGCCACGTGGATATTAAGAGTGAATTAGGTTACGAAGGTGGCCAAAAT
AGCGGTAATCAGTCATTTGAGGAAGACACAGAAGAAGATAAACCGAAATATGAACAAGGT
GGCAATATCGTAGATATCGATTTCGATAGTGTACCTCAAATTCATGGTCAAAATAATGGT
AACCAATCATTCGAAGAAGATACAGAGAAAGACAAACCTAAGTATGAACAAGGTGGTAAT
ATCATTGATATCGACTTCGACAGTGTGCCACATATTCACGGATTCAATAAGCACACTGAA
ATTATTGAAGAAGATACAAATAAAGATAAACCAAATTATCAATTCGGTGGACACAATAGT
GTTGACTTTGAAGAAGATACACTTCCACAAGTAAGTGGTCATAATGAAGGTCAACAAACG
ATTGAAGAAGATACAACACCTCCAATCGTGCCACCAACGCCACCGACACCAGAAGTACCA
AGCGAGCCGGAAACACCAACACCACCGACACCAGAAGTACCAAGCGAGCCGGAAACACCA
ACACCGCCAACGCCAGAGGTACCAACTGAACCTGGTAAACCAATACCACCTGCTAAAGAA
GAACCTAAAAAACCTTCTAAACCAGTGGAACAAGGTAAAGTAGTAACACCTGTTATTGAA
ATCAATGAAAAGGTTAAAGCAGTGGTACCAACTAAAAAAGCACAATCTAAGAAATCTGAA
CTACCTGAAACAGGTGGAGAAGAATCAACAAACAACGGCATGTTGTTCGGCGGATTATTT
AGCATTTTAGGTTTAGCGTTATTACGCAGAAATAAAAAGAATCACAAAGCATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2754
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02802
- symbol: SAOUHSC_02802
- description: fibronectin binding protein B
- length: 917
- theoretical pI: 4.29721
- theoretical MW: 101024
- GRAVY: -0.888877
⊟Function[edit | edit source]
- TIGRFAM: Cell envelope Other LPXTG cell wall anchor domain (TIGR01167; HMM-score: 26.5)
- TheSEED :
- Fibronectin binding protein FnbB
- PFAM: Adhesin (CL0204) SdrG_C_C; C-terminus of bacterial fibrinogen-binding adhesin (PF10425; HMM-score: 133.5)no clan defined Fn_bind; Fibronectin binding repeat (PF02986; HMM-score: 107.8)and 1 moreGram_pos_anchor; LPXTG cell wall anchor motif (PF00746; HMM-score: 34.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 10
- Extracellular Score: 0
- Internal Helices: 0
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 0.83
- Signal peptide possibility: -1
- N-terminally Anchored Score: -2
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.063431
- TAT(Tat/SPI): 0.01124
- LIPO(Sec/SPII): 0.01857
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIVVGMGQEKEAAASEQNNTTVEESGSSATESKASETQTTTNNVNTIDETQSYSATSTEQPSQSTQVTTEEAPKTVQAPKVETSRVDLPSEKVADKETTGTQVDIAQPSNVSEIKPRMKRSTDVTAVAEKEVVEETKATGTDVTNKVEVEEGSEIVGHKQDTNVVNPHNAERVTLKYKWKFGEGIKAGDYFDFTLSDNVETHGISTLRKVPEIKSTDGQVMATGEIIGERKVRYTFKEYVQEKKDLTAELSLNLFIDPTTVTQKGNQNVEVKLGETTVSKIFNIQYLGGVRDNWGVTANGRIDTLNKVDGKFSHFAYMKPNNQSLSSVTVTGQVTKGNKPGVNNPTVKVYKHIGSDDLAESVYAKLDDVSKFEDVTDNMSLDFDTNGGYSLNFNNLDQSKNYVIKYEGYYDSNASNLEFQTHLFGYYNYYYTSNLTWKNGVAFYSNNAQGDGKDKLKEPIIEHSTPIELEFKSEPPVEKHELTGTIEESNDSKPIDFEYHTAVEGAEGHAEGTIETEEDSIHVDFEESTHENSKHHADVVEYEEDTNPGGGQVTTESNLVEFDEDSTKGIVTGAVSDHTTIEDTKEYTTESNLIELVDELPEEHGQAQGPIEEITENNHHISHSGLGTENGHGNYGVIEEIEENSHVDIKSELGYEGGQNSGNQSFEEDTEEDKPKYEQGGNIVDIDFDSVPQIHGQNNGNQSFEEDTEKDKPKYEQGGNIIDIDFDSVPHIHGFNKHTEIIEEDTNKDKPNYQFGGHNSVDFEEDTLPQVSGHNEGQQTIEEDTTPPIVPPTPPTPEVPSEPETPTPPTPEVPSEPETPTPPTPEVPTEPGKPIPPAKEEPKKPSKPVEQGKVVTPVIEINEKVKAVVPTKKAQSKKSELPETGGEESTNNGMLFGGLFSILGLALLRRNKKNHKA
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
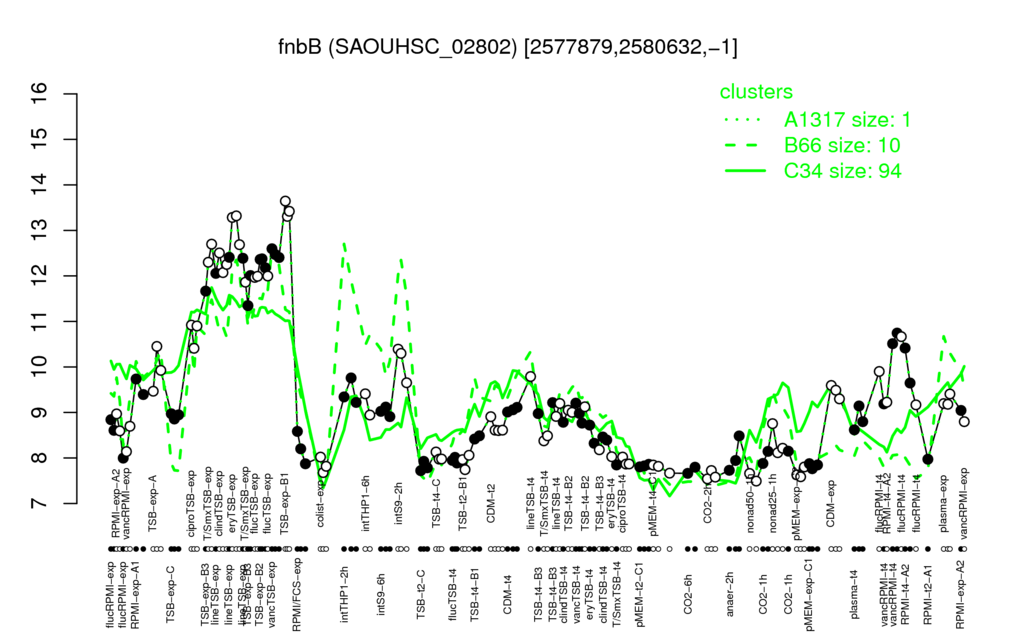 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You are kindly invited to share additional interesting facts.
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
T J Foster, M Höök
Surface protein adhesins of Staphylococcus aureus.
Trends Microbiol: 1998, 6(12);484-8
[PubMed:10036727] [WorldCat.org] [DOI] (P p)
