Jump to navigation
Jump to search
m (Text replacement - "gene Genbank" to "gene RefSeq") |
m (Text replacement - "* <aureodatabase>protein Genbank</aureodatabase> " to "") |
||
| Line 1: | Line 1: | ||
__TOC__ | |||
<protect> | <protect> | ||
<aureodatabase> | <aureodatabase>annotation</aureodatabase> | ||
=Summary= | =Summary= | ||
* <aureodatabase>organism</aureodatabase> | *<aureodatabase>organism</aureodatabase> | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>pan locus</aureodatabase> | *<aureodatabase>pan locus</aureodatabase> | ||
* <aureodatabase>gene symbol</aureodatabase> | *<aureodatabase>gene symbol</aureodatabase> | ||
* <aureodatabase>pan gene symbol</aureodatabase> | *<aureodatabase>pan gene symbol</aureodatabase> | ||
* <aureodatabase>gene synonyms</aureodatabase> | *<aureodatabase>gene synonyms</aureodatabase> | ||
* <aureodatabase>product</aureodatabase> | *<aureodatabase>product</aureodatabase> | ||
</protect> | </protect> | ||
| Line 24: | Line 25: | ||
==General== | ==General== | ||
* <aureodatabase>gene type</aureodatabase> | *<aureodatabase>gene type</aureodatabase> | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>gene symbol</aureodatabase> | *<aureodatabase>gene symbol</aureodatabase> | ||
* <aureodatabase>product</aureodatabase> | *<aureodatabase>product</aureodatabase> | ||
* <aureodatabase>gene replicon</aureodatabase> | *<aureodatabase>gene replicon</aureodatabase> | ||
* <aureodatabase>strand</aureodatabase> | *<aureodatabase>strand</aureodatabase> | ||
* <aureodatabase>gene coordinates</aureodatabase> | *<aureodatabase>gene coordinates</aureodatabase> | ||
* <aureodatabase>gene length</aureodatabase> | *<aureodatabase>gene length</aureodatabase> | ||
* <aureodatabase>essential</aureodatabase> | *<aureodatabase>essential</aureodatabase> | ||
*<aureodatabase>gene comment</aureodatabase> | |||
</protect> | </protect> | ||
| Line 38: | Line 40: | ||
==Accession numbers== | ==Accession numbers== | ||
* <aureodatabase>gene GI</aureodatabase> | *<aureodatabase>gene GI</aureodatabase> | ||
* <aureodatabase>gene RefSeq</aureodatabase> | *<aureodatabase>gene RefSeq</aureodatabase> | ||
*<aureodatabase>gene BioCyc</aureodatabase> | |||
*<aureodatabase>gene MicrobesOnline</aureodatabase> | |||
</protect> | </protect> | ||
<protect> | <protect> | ||
==Phenotype== | ==Phenotype== | ||
</protect> | </protect> | ||
Share your knowledge and add information here. [<span class="plainlinks">[//aureowiki.med.uni-greifswald.de/index.php?title={{PAGENAMEE}}&veaction=edit§ion=6 edit]</span>] | |||
<protect> | <protect> | ||
==DNA sequence== | ==DNA sequence== | ||
* <aureodatabase>gene sequence</aureodatabase> | *<aureodatabase>gene sequence</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
<aureodatabase>RNA regulated operons</aureodatabase> | |||
</protect> | |||
<protect> | |||
=Protein= | =Protein= | ||
<aureodatabase>protein 3D view</aureodatabase> | <aureodatabase>protein 3D view</aureodatabase> | ||
==General== | ==General== | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>protein symbol</aureodatabase> | *<aureodatabase>protein symbol</aureodatabase> | ||
* <aureodatabase>protein description</aureodatabase> | *<aureodatabase>protein description</aureodatabase> | ||
* <aureodatabase>protein length</aureodatabase> | *<aureodatabase>protein length</aureodatabase> | ||
* <aureodatabase>theoretical pI</aureodatabase> | *<aureodatabase>theoretical pI</aureodatabase> | ||
* <aureodatabase>theoretical MW</aureodatabase> | *<aureodatabase>theoretical MW</aureodatabase> | ||
* <aureodatabase>GRAVY</aureodatabase> | *<aureodatabase>GRAVY</aureodatabase> | ||
</protect> | </protect> | ||
| Line 71: | Line 78: | ||
==Function== | ==Function== | ||
* <aureodatabase>protein reaction</aureodatabase> | *<aureodatabase>protein reaction</aureodatabase> | ||
* <aureodatabase>protein TIGRFAM</aureodatabase> | *<aureodatabase>protein TIGRFAM</aureodatabase> | ||
* <aureodatabase>protein TheSeed</aureodatabase> | *<aureodatabase>protein TheSeed</aureodatabase> | ||
* <aureodatabase>protein PFAM</aureodatabase> | *<aureodatabase>protein PFAM</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
==Structure, modifications & | ==Structure, modifications & cofactors== | ||
* <aureodatabase>protein domains</aureodatabase> | *<aureodatabase>protein domains</aureodatabase> | ||
* <aureodatabase>protein modifications</aureodatabase> | *<aureodatabase>protein modifications</aureodatabase> | ||
* <aureodatabase>protein cofactors</aureodatabase> | *<aureodatabase>protein cofactors</aureodatabase> | ||
* <aureodatabase>protein effectors</aureodatabase> | *<aureodatabase>protein effectors</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein regulated operons</aureodatabase> | ||
</protect> | </protect> | ||
| Line 90: | Line 97: | ||
==Localization== | ==Localization== | ||
* <aureodatabase>protein Psortb</aureodatabase> | *<aureodatabase>protein Psortb</aureodatabase> | ||
* <aureodatabase>protein LocateP</aureodatabase> | *<aureodatabase>protein LocateP</aureodatabase> | ||
* <aureodatabase>protein SignalP</aureodatabase> | *<aureodatabase>protein SignalP</aureodatabase> | ||
* <aureodatabase>protein TMHMM</aureodatabase> | *<aureodatabase>protein TMHMM</aureodatabase> | ||
</protect> | </protect> | ||
| Line 99: | Line 106: | ||
==Accession numbers== | ==Accession numbers== | ||
* <aureodatabase>protein GI</aureodatabase> | *<aureodatabase>protein GI</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein RefSeq</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein UniProt</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein STRING</aureodatabase> | ||
</protect> | </protect> | ||
| Line 108: | Line 115: | ||
==Protein sequence== | ==Protein sequence== | ||
* <aureodatabase>protein sequence</aureodatabase> | *<aureodatabase>protein sequence</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
== | ==Experimental data== | ||
* <aureodatabase>protein validated peptides</aureodatabase> | *<aureodatabase>protein validated peptides</aureodatabase> | ||
*<aureodatabase>protein validated localization</aureodatabase> | |||
*<aureodatabase>protein validated quantitative data</aureodatabase> | |||
*<aureodatabase>protein partners</aureodatabase> | |||
</protect> | </protect> | ||
| Line 125: | Line 135: | ||
==Operon== | ==Operon== | ||
* <aureodatabase>operons</aureodatabase> | *<aureodatabase>operons</aureodatabase> | ||
</protect> | </protect> | ||
| Line 131: | Line 141: | ||
==Regulation== | ==Regulation== | ||
*<aureodatabase>regulators</aureodatabase> | |||
* <aureodatabase>regulators</aureodatabase> | |||
</protect> | </protect> | ||
| Line 138: | Line 147: | ||
==Transcription pattern== | ==Transcription pattern== | ||
* <aureodatabase>expression browser</aureodatabase> | *<aureodatabase>expression browser</aureodatabase> | ||
</protect> | </protect> | ||
| Line 144: | Line 153: | ||
==Protein synthesis (provided by Aureolib)== | ==Protein synthesis (provided by Aureolib)== | ||
* <aureodatabase>protein synthesis Aureolib</aureodatabase> | *<aureodatabase>protein synthesis Aureolib</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
== | ==Protein stability== | ||
* <aureodatabase>protein half-life</aureodatabase> | *<aureodatabase>protein half-life</aureodatabase> | ||
</protect> | </protect> | ||
Latest revision as of 23:05, 10 March 2016
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02274
- pan locus tag?: SAUPAN005292000
- symbol: SAOUHSC_02274
- pan gene symbol?: vga
- synonym:
- product: ABC transporter ATP-binding protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02274
- symbol: SAOUHSC_02274
- product: ABC transporter ATP-binding protein
- replicon: chromosome
- strand: +
- coordinates: 2104734..2106662
- length: 1929
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919149 NCBI
- RefSeq: YP_500755 NCBI
- BioCyc: G1I0R-2145 BioCyc
- MicrobesOnline: 1290711 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921ATGATACTTTTACAACTCAATCATATATCAAAATCGTTCGATGGTGAAGATATATTTACT
GATGTTGATTTTGAAGTAAAAACTGGTGAACGAATTGGTATCGTAGGAAGAAATGGCGCC
GGTAAATCAACATTGATGAAAATTATAGCTGGCGTAGAAAACTATGATTCTGGAAATGTG
TCAAAAATTAAAAACTTAAAACTAGGTTATTTAACTCAACAAATGACTTTAAATTCTAAT
GCAACAGTTTTTGAAGAAATGTCAAAACCATTTGAACATATTAAACGAATGGAAAGCTTA
ATCAAAGAAGAAACAGATTGGTTATCAAAACATGCAAATGATTACGATAGTGATACATAT
AAAACACATATGTCCCGTTATGAATCTTTATCGAATCAATTTGAACAATTAGAAGGATAT
CAATATGAAAGTAAAATTAAAACAGTACTTTACGGGTTAAATTTTAGTGAAGAAGATTTC
AATAAACCTATCAATGATTTTAGCGGTGGCCAAAAAACACGTTTATCTTTAGCTCAAATG
CTATTAAACGAACCTGATTTATTACTTTTAGATGAACCTACTAACCACTTAGATTTAGAA
ACGACAAAGTGGCTTGAAGATTATCTACGTTATTTTAAAGGTGCAATCGTCATCATCAGC
CATGATCGTTACTTTTTAGATAAAATAGTTACTCAAATTTATGATGTGGCTTTAGGTGAT
GTCAAACGCTATGTTGGTAATTACGAGGAATTTATACAGCAACGGGATTTATATTATCAA
AAACGAATGCAAGAATATGAAAGTCAACAAGCAGAAATAAAACGATTAGAAACTTTTGTT
GAGAAAAATATTACCCGTGCTTCAACAAGTGGAATGGCAAAAAGTAGACGTAAGATTTTA
GAAAAAATGGAACGCATTGATAAACCAATGTTAGATGCCAAAAGTGCAAATATTCAATTT
GGCTTTGACCGGAATACAGGTAATGACGTCATGCATGTAAAAAATTTAGAAATCGGTTAT
CAAACTGCAATTACCAAACCTATGAGTATAGAGGTCTCTAAAGGCGATCATATAGCAATC
ATTGGGCCAAATGGTATTGGAAAATCGACCTTAATTAAAACTATTGCTAATCAACAAAAA
GCGCTTAATGGCGATATTACTTTCGGCGCAAATTTACAAATTGGTTATTATGATCAAAAG
CAAGCAGAATTTAAATCTAGTAAAACGATTTTAGATTATGTGTGGGATCAATATCCGTTA
ATGAATGAAAAAGATATTCGAGCAGTTCTTGGACGTTTCTTATTTGTACAAGACGATGTT
AAAAAGATAATTAATGATTTATCTGGTGGTGAAAAAGCACGTTTACAACTAGCACTACTT
ATGTTGCAACGTGACAATGTACTTATTTTAGATGAACCTACCAACCACCTCGATATAGAT
TCAAAAGAAATGTTAGAGCAAGCACTCCAACATTTTGAAGGAACAATACTATTTGTTTCC
CATGATCGTTACTTTATTAACCAATTAGCAAACAAGGTATTTGATTTAACAATTGATGGC
GGAAAGATGTATTTAGGAGATTATCAATATTACATTGAAAAAGTTGAGGAAGCGGAAGCG
TTGAAAGCGCACCAAGAAGAACAGTCAGTTAACGTACAAGCACATAAAAAGTCTATGGAA
CAATCGTCGTATCATAACCAAAAAGAACAAAGACGTGAACAACGCAAACTTGAACGACAA
ATATCCGAGTGTGAAAATGAAATAGAAACTTTAGAGACTACTATCTTACAAATAGACGAG
CAATTAACCCAACCAGAAGTGTATAACAATCCACAAAAAGCAAACGAATTAGCTATCCAA
AAACAAGATAGCGAACAAAAATTAGAACACGCTATGTCTAAATGGGAAGAATTACAACAA
AAACTATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1929
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02274
- symbol: SAOUHSC_02274
- description: ABC transporter ATP-binding protein
- length: 642
- theoretical pI: 5.07123
- theoretical MW: 74460.6
- GRAVY: -0.649844
⊟Function[edit | edit source]
- TIGRFAM: ATP-binding cassette protein, ChvD family (TIGR03719; HMM-score: 392)and 94 moreCellular processes Toxin production and resistance putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 172.9)Transport and binding proteins Unknown substrate putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 172.9)thiol reductant ABC exporter, CydD subunit (TIGR02857; HMM-score: 160.2)thiol reductant ABC exporter, CydC subunit (TIGR02868; HMM-score: 150.5)Protein fate Protein and peptide secretion and trafficking heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 148.8)Transport and binding proteins Other heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 148.8)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtD (TIGR03411; HMM-score: 144.6)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04521; EC 3.6.3.-; HMM-score: 143.4)Transport and binding proteins Anions phosphonate ABC transporter, ATP-binding protein (TIGR02315; EC 3.6.3.28; HMM-score: 141.8)proposed F420-0 ABC transporter, ATP-binding protein (TIGR03873; HMM-score: 141)Cellular processes Pathogenesis type I secretion system ATPase (TIGR03375; HMM-score: 139.8)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR03375; HMM-score: 139.8)Cellular processes Cell division cell division ATP-binding protein FtsE (TIGR02673; HMM-score: 136.7)lantibiotic protection ABC transporter, ATP-binding subunit (TIGR03740; HMM-score: 135.7)Transport and binding proteins Other daunorubicin resistance ABC transporter, ATP-binding protein (TIGR01188; HMM-score: 135.6)Protein fate Protein and peptide secretion and trafficking lipoprotein releasing system, ATP-binding protein (TIGR02211; EC 3.6.3.-; HMM-score: 135.2)Biosynthesis of cofactors, prosthetic groups, and carriers Other FeS assembly ATPase SufC (TIGR01978; HMM-score: 133.5)Transport and binding proteins Carbohydrates, organic alcohols, and acids ABC transporter, ATP-binding subunit, PQQ-dependent alcohol dehydrogenase system (TIGR03864; HMM-score: 131.7)Transport and binding proteins Cations and iron carrying compounds cobalt ABC transporter, ATP-binding protein (TIGR01166; HMM-score: 129.4)Transport and binding proteins Anions sulfate ABC transporter, ATP-binding protein (TIGR00968; EC 3.6.3.25; HMM-score: 128.3)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 128.3)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 128.3)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04520; EC 3.6.3.-; HMM-score: 127.5)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtE (TIGR03410; HMM-score: 126.3)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 122.7)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 122.7)Transport and binding proteins Anions molybdate ABC transporter, ATP-binding protein (TIGR02142; EC 3.6.3.29; HMM-score: 120.5)Transport and binding proteins Amino acids, peptides and amines putative 2-aminoethylphosphonate ABC transporter, ATP-binding protein (TIGR03265; HMM-score: 120.3)Energy metabolism Methanogenesis methyl coenzyme M reductase system, component A2 (TIGR03269; HMM-score: 117.3)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01842; HMM-score: 117.2)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikE (TIGR02769; EC 3.6.3.24; HMM-score: 114.4)gliding motility-associated ABC transporter ATP-binding subunit GldA (TIGR03522; HMM-score: 113.1)Transport and binding proteins Anions phosphate ABC transporter, ATP-binding protein (TIGR00972; EC 3.6.3.27; HMM-score: 112.8)ABC exporter ATP-binding subunit, DevA family (TIGR02982; HMM-score: 112.3)Cellular processes Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 112.2)Transport and binding proteins Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 112.2)Transport and binding proteins Other thiamine ABC transporter, ATP-binding protein (TIGR01277; EC 3.6.3.-; HMM-score: 111.9)Protein fate Protein and peptide secretion and trafficking ABC-type bacteriocin transporter (TIGR01193; HMM-score: 108.5)Protein fate Protein modification and repair ABC-type bacteriocin transporter (TIGR01193; HMM-score: 108.5)Transport and binding proteins Other ABC-type bacteriocin transporter (TIGR01193; HMM-score: 108.5)phosphonate C-P lyase system protein PhnL (TIGR02324; HMM-score: 108.2)Transport and binding proteins Unknown substrate anchored repeat-type ABC transporter, ATP-binding subunit (TIGR03771; HMM-score: 101.7)Transport and binding proteins Carbohydrates, organic alcohols, and acids D-xylose ABC transporter, ATP-binding protein (TIGR02633; EC 3.6.3.17; HMM-score: 100.7)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 100.6)Transport and binding proteins Other lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 100.6)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 100.4)Transport and binding proteins Other LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 100.4)2-aminoethylphosphonate ABC transport system, ATP-binding component PhnT (TIGR03258; HMM-score: 99.2)Transport and binding proteins Anions nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 98.8)Transport and binding proteins Other nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 98.8)Transport and binding proteins Other antigen peptide transporter 2 (TIGR00958; HMM-score: 98.3)ABC transporter, permease/ATP-binding protein (TIGR02204; HMM-score: 97.3)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01846; HMM-score: 95.8)ectoine/hydroxyectoine ABC transporter, ATP-binding protein EhuA (TIGR03005; HMM-score: 88.1)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikD (TIGR02770; HMM-score: 87.6)Transport and binding proteins Other pigment precursor permease (TIGR00955; HMM-score: 87.1)Transport and binding proteins Carbohydrates, organic alcohols, and acids glucan exporter ATP-binding protein (TIGR01192; EC 3.6.3.42; HMM-score: 85.3)Transport and binding proteins Carbohydrates, organic alcohols, and acids peroxysomal long chain fatty acyl transporter (TIGR00954; HMM-score: 84.8)Transport and binding proteins Amino acids, peptides and amines polyamine ABC transporter, ATP-binding protein (TIGR01187; EC 3.6.3.31; HMM-score: 82.2)Transport and binding proteins Amino acids, peptides and amines glycine betaine/L-proline transport ATP binding subunit (TIGR01186; HMM-score: 78.9)D-methionine ABC transporter, ATP-binding protein (TIGR02314; EC 3.6.3.-; HMM-score: 77)Central intermediary metabolism Phosphorus compounds phosphonate C-P lyase system protein PhnK (TIGR02323; HMM-score: 74.1)Transport and binding proteins Anions cystic fibrosis transmembrane conductor regulator (CFTR) (TIGR01271; EC 3.6.3.49; HMM-score: 67.9)Transport and binding proteins Amino acids, peptides and amines cyclic peptide transporter (TIGR01194; HMM-score: 59.2)Transport and binding proteins Other cyclic peptide transporter (TIGR01194; HMM-score: 59.2)Transport and binding proteins Amino acids, peptides and amines choline ABC transporter, ATP-binding protein (TIGR03415; HMM-score: 57.8)Transport and binding proteins Other pleiotropic drug resistance family protein (TIGR00956; HMM-score: 51)Transport and binding proteins Other multi drug resistance-associated protein (MRP) (TIGR00957; HMM-score: 50.7)Transport and binding proteins Other rim ABC transporter (TIGR01257; HMM-score: 38.1)Cellular processes Chemotaxis and motility flagellar protein export ATPase FliI (TIGR03496; EC 3.6.3.14; HMM-score: 26.6)Cellular processes Pathogenesis type III secretion apparatus H+-transporting two-sector ATPase (TIGR02546; EC 3.6.3.14; HMM-score: 25.9)Protein fate Protein and peptide secretion and trafficking type III secretion apparatus H+-transporting two-sector ATPase (TIGR02546; EC 3.6.3.14; HMM-score: 25.9)Protein synthesis Other ribosome-associated GTPase EngA (TIGR03594; HMM-score: 24.5)Energy metabolism ATP-proton motive force interconversion ATPase, FliI/YscN family (TIGR01026; EC 3.6.3.14; HMM-score: 24.3)Unknown function General small GTP-binding protein domain (TIGR00231; HMM-score: 24.1)Cellular processes Chemotaxis and motility flagellar protein export ATPase FliI (TIGR03497; EC 3.6.3.14; HMM-score: 24.1)flagellar protein export ATPase FliI (TIGR03498; EC 3.6.3.14; HMM-score: 23.3)Protein synthesis Other GTP-binding protein Era (TIGR00436; HMM-score: 21.2)Protein synthesis Other ribosome biogenesis GTP-binding protein YsxC (TIGR03598; HMM-score: 19.1)DNA metabolism DNA replication, recombination, and repair Holliday junction DNA helicase RuvB (TIGR00635; EC 3.6.4.12; HMM-score: 18.1)Purines, pyrimidines, nucleosides, and nucleotides Nucleotide and nucleoside interconversions cytidylate kinase (TIGR00017; EC 2.7.4.14; HMM-score: 17.9)Protein synthesis Other Obg family GTPase CgtA (TIGR02729; HMM-score: 17.9)Central intermediary metabolism Phosphorus compounds phosphonate metabolism protein/1,5-bisphosphokinase (PRPP-forming) PhnN (TIGR02322; HMM-score: 15.1)Biosynthesis of cofactors, prosthetic groups, and carriers Pantothenate and coenzyme A dephospho-CoA kinase (TIGR00152; EC 2.7.1.24; HMM-score: 15)Protein fate Protein and peptide secretion and trafficking type VII secretion protein EccCb (TIGR03925; HMM-score: 14.9)Protein synthesis Translation factors ribosome small subunit-dependent GTPase A (TIGR00157; EC 3.6.-.-; HMM-score: 14.1)Protein fate Protein modification and repair [FeFe] hydrogenase H-cluster maturation GTPase HydF (TIGR03918; HMM-score: 10.9)Transport and binding proteins Amino acids, peptides and amines LAO/AO transport system ATPase (TIGR00750; EC 2.7.-.-; HMM-score: 10.8)Regulatory functions Protein interactions LAO/AO transport system ATPase (TIGR00750; EC 2.7.-.-; HMM-score: 10.8)Central intermediary metabolism Nitrogen metabolism urea carboxylase (TIGR02712; EC 6.3.4.6; HMM-score: 9.9)putative cytidylate kinase (TIGR02173; EC 2.7.4.14; HMM-score: 9.5)Cellular processes Chemotaxis and motility flagellar biosynthesis protein FlhF (TIGR03499; HMM-score: 9.1)Cellular processes Cell division chromosome segregation protein SMC (TIGR02168; HMM-score: 4)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02168; HMM-score: 4)
- TheSEED :
- ABC transporter ATP-binding protein uup
- PFAM: P-loop_NTPase (CL0023) ABC_tran; ABC transporter (PF00005; HMM-score: 169.6)and 42 moreABC_tran_Xtn; ABC transporter (PF12848; HMM-score: 76.9)AAA_21; AAA domain, putative AbiEii toxin, Type IV TA system (PF13304; HMM-score: 75.1)no clan defined ABC_tran_CTD; ABC transporter C-terminal domain (PF16326; HMM-score: 47.4)P-loop_NTPase (CL0023) SMC_N; RecF/RecN/SMC N terminal domain (PF02463; HMM-score: 44.8)AAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 34.4)MMR_HSR1; 50S ribosome-binding GTPase (PF01926; HMM-score: 30.3)AAA_22; AAA domain (PF13401; HMM-score: 28.4)NB-ARC; NB-ARC domain (PF00931; HMM-score: 26.6)RsgA_GTPase; RsgA GTPase (PF03193; HMM-score: 26.2)AAA_24; AAA domain (PF13479; HMM-score: 25.4)NACHT; NACHT domain (PF05729; HMM-score: 24.1)AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 23.9)ATP-synt_ab; ATP synthase alpha/beta family, nucleotide-binding domain (PF00006; HMM-score: 22.8)IstB_IS21; IstB-like ATP binding protein (PF01695; HMM-score: 22.5)Roc; Ras of Complex, Roc, domain of DAPkinase (PF08477; HMM-score: 22.2)RNA_helicase; RNA helicase (PF00910; HMM-score: 21.4)ATPase_2; ATPase domain predominantly from Archaea (PF01637; HMM-score: 20.8)AAA_16; AAA ATPase domain (PF13191; HMM-score: 20.6)NTPase_1; NTPase (PF03266; HMM-score: 19.2)AAA_30; AAA domain (PF13604; HMM-score: 19.2)AAA_10; AAA-like domain (PF12846; HMM-score: 18.9)AAA_28; AAA domain (PF13521; HMM-score: 18.7)AAA_18; AAA domain (PF13238; HMM-score: 18.6)Dynamin_N; Dynamin family (PF00350; HMM-score: 17.7)AAA_33; AAA domain (PF13671; HMM-score: 17.5)FtsK_SpoIIIE; FtsK/SpoIIIE family (PF01580; HMM-score: 16.1)no clan defined SL4P; Uncharacterized Strongylid L4 protein (PF17618; HMM-score: 16)P-loop_NTPase (CL0023) Cytidylate_kin; Cytidylate kinase (PF02224; HMM-score: 15.8)TsaE; Threonylcarbamoyl adenosine biosynthesis protein TsaE (PF02367; HMM-score: 15.2)ArgK; ArgK protein (PF03308; HMM-score: 14.6)AAA_14; AAA domain (PF13173; HMM-score: 14.5)HAD (CL0137) Trehalose_PPase; Trehalose-phosphatase (PF02358; HMM-score: 14.2)P-loop_NTPase (CL0023) RuvB_N; Holliday junction DNA helicase ruvB N-terminus (PF05496; HMM-score: 14)cobW; CobW/HypB/UreG, nucleotide-binding domain (PF02492; HMM-score: 13.1)AAA_5; AAA domain (dynein-related subfamily) (PF07728; HMM-score: 12.5)Mg_chelatase; Magnesium chelatase, subunit ChlI (PF01078; HMM-score: 11.6)DUF87; Domain of unknown function DUF87 (PF01935; HMM-score: 11.4)AAA_27; AAA domain (PF13514; HMM-score: 11)C_Lectin (CL0056) Endostatin; Collagenase NC10 and Endostatin (PF06482; HMM-score: 8.5)P-loop_NTPase (CL0023) AAA_23; AAA domain (PF13476; HMM-score: 8.3)SRP54; SRP54-type protein, GTPase domain (PF00448; HMM-score: 6.8)no clan defined DASH_Spc34; DASH complex subunit Spc34 (PF08657; HMM-score: 6.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.021608
- TAT(Tat/SPI): 0.00242
- LIPO(Sec/SPII): 0.000955
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MILLQLNHISKSFDGEDIFTDVDFEVKTGERIGIVGRNGAGKSTLMKIIAGVENYDSGNVSKIKNLKLGYLTQQMTLNSNATVFEEMSKPFEHIKRMESLIKEETDWLSKHANDYDSDTYKTHMSRYESLSNQFEQLEGYQYESKIKTVLYGLNFSEEDFNKPINDFSGGQKTRLSLAQMLLNEPDLLLLDEPTNHLDLETTKWLEDYLRYFKGAIVIISHDRYFLDKIVTQIYDVALGDVKRYVGNYEEFIQQRDLYYQKRMQEYESQQAEIKRLETFVEKNITRASTSGMAKSRRKILEKMERIDKPMLDAKSANIQFGFDRNTGNDVMHVKNLEIGYQTAITKPMSIEVSKGDHIAIIGPNGIGKSTLIKTIANQQKALNGDITFGANLQIGYYDQKQAEFKSSKTILDYVWDQYPLMNEKDIRAVLGRFLFVQDDVKKIINDLSGGEKARLQLALLMLQRDNVLILDEPTNHLDIDSKEMLEQALQHFEGTILFVSHDRYFINQLANKVFDLTIDGGKMYLGDYQYYIEKVEEAEALKAHQEEQSVNVQAHKKSMEQSSYHNQKEQRREQRKLERQISECENEIETLETTILQIDEQLTQPEVYNNPQKANELAIQKQDSEQKLEHAMSKWEELQQKL
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
SAOUHSC_01246 (infB) translation initiation factor IF-2 [3] (data from MRSA252) SAOUHSC_00900 (pgi) glucose-6-phosphate isomerase [3] (data from MRSA252) SAOUHSC_00796 (pgk) phosphoglycerate kinase [3] (data from MRSA252) SAOUHSC_02511 (rplD) 50S ribosomal protein L4 [3] (data from MRSA252) SAOUHSC_02496 (rplF) 50S ribosomal protein L6 [3] (data from MRSA252) SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [3] (data from MRSA252) SAOUHSC_01211 (rplS) 50S ribosomal protein L19 [3] (data from MRSA252) SAOUHSC_00524 (rpoB) DNA-directed RNA polymerase subunit beta [3] (data from MRSA252) SAOUHSC_01232 (rpsB) 30S ribosomal protein S2 [3] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_02487 (rpsM) 30S ribosomal protein S13 [3] (data from MRSA252) SAOUHSC_02503 (rpsQ) 30S ribosomal protein S17 [3] (data from MRSA252) SAOUHSC_00284 5'-nucleotidase [3] (data from MRSA252) SAOUHSC_00529 elongation factor G [3] (data from MRSA252) SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_00795 glyceraldehyde-3-phosphate dehydrogenase [3] (data from MRSA252) SAOUHSC_00878 hypothetical protein [3] (data from MRSA252) SAOUHSC_01040 pyruvate dehydrogenase complex, E1 component subunit alpha [3] (data from MRSA252) SAOUHSC_01150 cell division protein FtsZ [3] (data from MRSA252) SAOUHSC_01218 succinyl-CoA synthetase subunit alpha [3] (data from MRSA252) SAOUHSC_01605 6-phosphogluconate dehydrogenase [3] (data from MRSA252) SAOUHSC_01666 glycyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01794 glyceraldehyde 3-phosphate dehydrogenase 2 [3] (data from MRSA252) SAOUHSC_01801 isocitrate dehydrogenase [3] (data from MRSA252) SAOUHSC_01806 pyruvate kinase [3] (data from MRSA252) SAOUHSC_01819 hypothetical protein [3] (data from MRSA252) SAOUHSC_02366 fructose-bisphosphate aldolase [3] (data from MRSA252) SAOUHSC_02377 pyrimidine-nucleoside phosphorylase [3] (data from MRSA252) SAOUHSC_02441 alkaline shock protein 23 [3] (data from MRSA252) SAOUHSC_02512a 30S ribosomal protein S10 [3] (data from MRSA252) SAOUHSC_02926 fructose-1,6-bisphosphate aldolase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
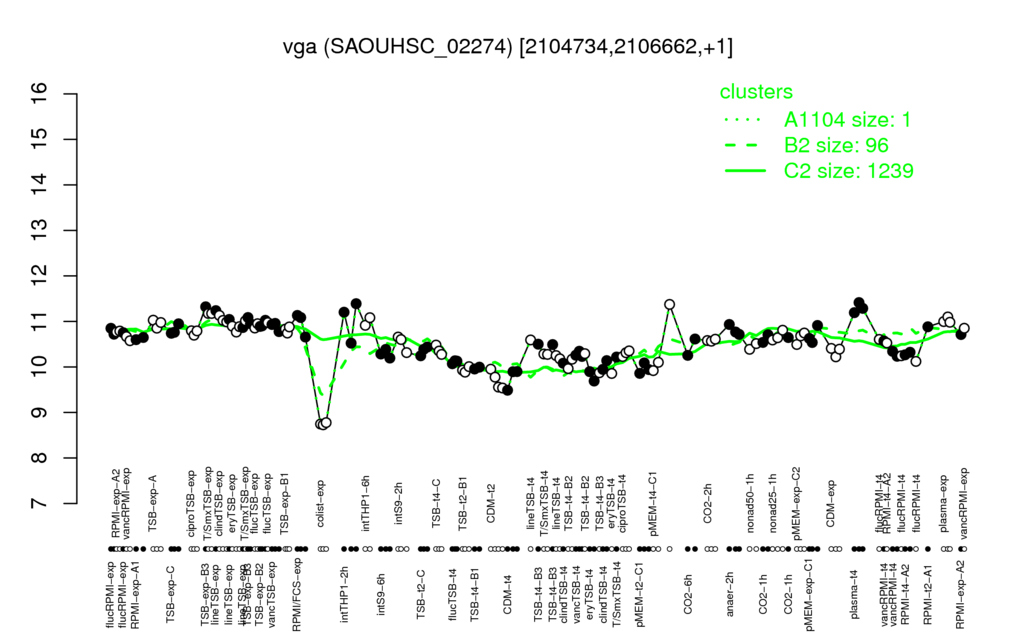 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You are kindly invited to share additional interesting facts.
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 3.24 3.25 3.26 3.27 3.28 3.29 3.30 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
