NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01897
- pan locus tag?: SAUPAN004470000
- symbol: SAOUHSC_01897
- pan gene symbol?: sigS
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01897
- symbol: SAOUHSC_01897
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 1808335..1808805
- length: 471
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920844 NCBI
- RefSeq: YP_500398 NCBI
- BioCyc: G1I0R-1763 BioCyc
- MicrobesOnline: 1290312 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421TTGAAATTTAATGACGTATACAACAAACACCAGAAAATCATACACTATCTTTTAAAAAAA
TATAATATTAGCTATAATTATGATGAGTATTATCAACTACTCTTGATAAAAATGTGGCAA
TTGAGTCAGATATATAAACCCTCAAGCAAGCAATCTTTATCCTCTTTTTTATTCACTCGA
TTAAATTTTTACCTTATCGATTTATTCAGACAACAAAATCAATTAAAAGATGTCATTTTA
TGTGAGAATAATTCACCAACATTAACTGAACAACCAACATACTTTAATGAACATGACCTT
CGTTTACAAGATATCTTCAAGCTTTTAAATCAAAGAGAAAGACTATGGCTCAAACTATAC
CTTGAAGGATACAAGCAATTTGAAATTGCTGAAATCATGTCATTATCGCTTTCAACGATT
AAATTAATTAAGATGTCCGTTAAGCGTAAATGCCAACATAATTTTAATTAG60
120
180
240
300
360
420
471
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01897
- symbol: SAOUHSC_01897
- description: hypothetical protein
- length: 156
- theoretical pI: 9.76636
- theoretical MW: 19153.2
- GRAVY: -0.469231
⊟Function[edit | edit source]
- TIGRFAM: RNA polymerase sigma factor, sigma-70 family (TIGR02937; HMM-score: 60.9)and 11 moretranscriptional regulator BotR, P-21 (TIGR03209; HMM-score: 28.8)RNA polymerase sigma factor, SigZ family (TIGR02959; HMM-score: 28)RNA polymerase sigma-70 factor, Bacteroides expansion family 1 (TIGR02985; HMM-score: 22.3)RNA polymerase sigma factor, FliA/WhiG family (TIGR02479; HMM-score: 21.8)RNA polymerase sigma-70 factor, TIGR02954 family (TIGR02954; HMM-score: 16.6)RNA polymerase sigma factor, TIGR02999 family (TIGR02999; HMM-score: 15)LuxR family transcriptional regulatory, chaperone HchA-associated (TIGR03541; HMM-score: 14.9)Cellular processes Sporulation and germination RNA polymerase sigma-G factor (TIGR02850; HMM-score: 12.4)Transcription Transcription factors RNA polymerase sigma-G factor (TIGR02850; HMM-score: 12.4)RNA polymerase sigma-B factor (TIGR02941; HMM-score: 12.1)RNA polymerase sigma-70 factor, sigma-E family (TIGR02983; HMM-score: 11.9)
- TheSEED :
- RNA polymerase sigma factor SigS
- PFAM: HTH (CL0123) Sigma70_r2; Sigma-70 region 2 (PF04542; HMM-score: 28.7)GerE; Bacterial regulatory proteins, luxR family (PF00196; HMM-score: 24.3)and 4 moreSigma70_r4; Sigma-70, region 4 (PF04545; HMM-score: 20.1)Sigma70_r4_2; Sigma-70, region 4 (PF08281; HMM-score: 19.4)Terminase_5; Putative ATPase subunit of terminase (gpP-like) (PF06056; HMM-score: 13.4)HTH_7; Helix-turn-helix domain of resolvase (PF02796; HMM-score: 12.9)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: unknown (no significant prediction)
- Cytoplasmic Score: 2.5
- Cytoplasmic Membrane Score: 2.5
- Cellwall Score: 2.5
- Extracellular Score: 2.5
- Internal Helices: 0
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003865
- TAT(Tat/SPI): 0.000202
- LIPO(Sec/SPII): 0.000979
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKFNDVYNKHQKIIHYLLKKYNISYNYDEYYQLLLIKMWQLSQIYKPSSKQSLSSFLFTRLNFYLIDLFRQQNQLKDVILCENNSPTLTEQPTYFNEHDLRLQDIFKLLNQRERLWLKLYLEGYKQFEIAEIMSLSLSTIKLIKMSVKRKCQHNFN
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
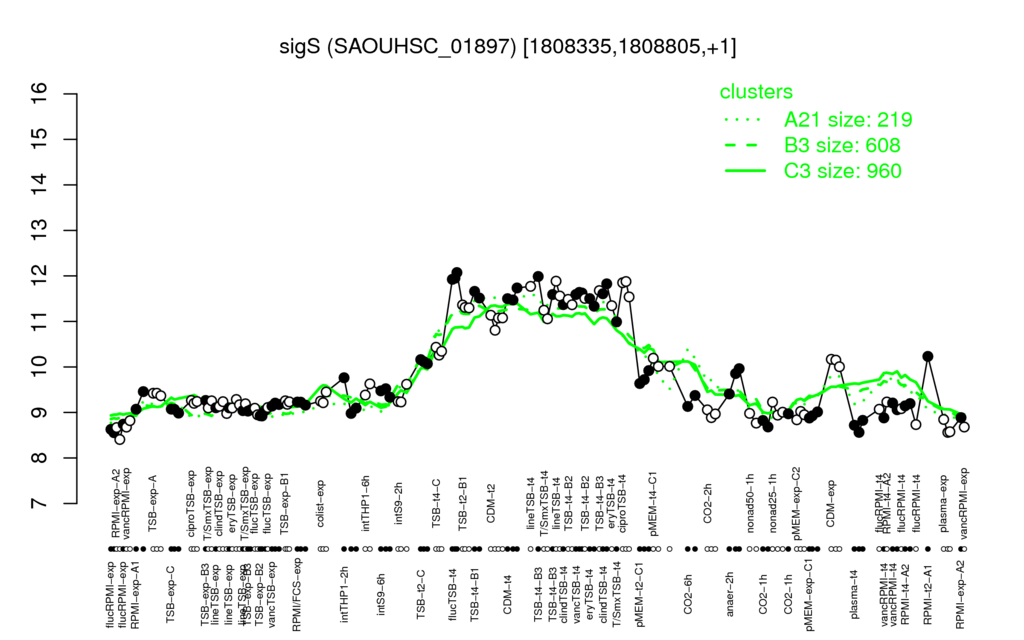 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You are kindly invited to share additional interesting facts.
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Lindsey N Shaw, Catharina Lindholm, Tomasz K Prajsnar, Halie K Miller, Melanie C Brown, Ewa Golonka, George C Stewart, Andrej Tarkowski, Jan Potempa
Identification and characterization of sigma, a novel component of the Staphylococcus aureus stress and virulence responses.
PLoS One: 2008, 3(12);e3844
[PubMed:19050758] [WorldCat.org] [DOI] (I p)
