Jump to navigation
Jump to search
m (Text replacement - "gene Genbank" to "gene RefSeq") |
m (Text replacement - "* <aureodatabase>protein Genbank</aureodatabase> " to "") |
||
| Line 1: | Line 1: | ||
__TOC__ | |||
<protect> | <protect> | ||
<aureodatabase> | <aureodatabase>annotation</aureodatabase> | ||
=Summary= | =Summary= | ||
* <aureodatabase>organism</aureodatabase> | *<aureodatabase>organism</aureodatabase> | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>pan locus</aureodatabase> | *<aureodatabase>pan locus</aureodatabase> | ||
* <aureodatabase>gene symbol</aureodatabase> | *<aureodatabase>gene symbol</aureodatabase> | ||
* <aureodatabase>pan gene symbol</aureodatabase> | *<aureodatabase>pan gene symbol</aureodatabase> | ||
* <aureodatabase>gene synonyms</aureodatabase> | *<aureodatabase>gene synonyms</aureodatabase> | ||
* <aureodatabase>product</aureodatabase> | *<aureodatabase>product</aureodatabase> | ||
</protect> | </protect> | ||
| Line 24: | Line 25: | ||
==General== | ==General== | ||
* <aureodatabase>gene type</aureodatabase> | *<aureodatabase>gene type</aureodatabase> | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>gene symbol</aureodatabase> | *<aureodatabase>gene symbol</aureodatabase> | ||
* <aureodatabase>product</aureodatabase> | *<aureodatabase>product</aureodatabase> | ||
* <aureodatabase>gene replicon</aureodatabase> | *<aureodatabase>gene replicon</aureodatabase> | ||
* <aureodatabase>strand</aureodatabase> | *<aureodatabase>strand</aureodatabase> | ||
* <aureodatabase>gene coordinates</aureodatabase> | *<aureodatabase>gene coordinates</aureodatabase> | ||
* <aureodatabase>gene length</aureodatabase> | *<aureodatabase>gene length</aureodatabase> | ||
* <aureodatabase>essential</aureodatabase> | *<aureodatabase>essential</aureodatabase> | ||
*<aureodatabase>gene comment</aureodatabase> | |||
</protect> | </protect> | ||
| Line 38: | Line 40: | ||
==Accession numbers== | ==Accession numbers== | ||
* <aureodatabase>gene GI</aureodatabase> | *<aureodatabase>gene GI</aureodatabase> | ||
* <aureodatabase>gene RefSeq</aureodatabase> | *<aureodatabase>gene RefSeq</aureodatabase> | ||
*<aureodatabase>gene BioCyc</aureodatabase> | |||
*<aureodatabase>gene MicrobesOnline</aureodatabase> | |||
</protect> | </protect> | ||
<protect> | <protect> | ||
==Phenotype== | ==Phenotype== | ||
</protect> | </protect> | ||
Share your knowledge and add information here. [<span class="plainlinks">[//aureowiki.med.uni-greifswald.de/index.php?title={{PAGENAMEE}}&veaction=edit§ion=6 edit]</span>] | |||
<protect> | <protect> | ||
==DNA sequence== | ==DNA sequence== | ||
* <aureodatabase>gene sequence</aureodatabase> | *<aureodatabase>gene sequence</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
<aureodatabase>RNA regulated operons</aureodatabase> | |||
</protect> | |||
<protect> | |||
=Protein= | =Protein= | ||
<aureodatabase>protein 3D view</aureodatabase> | <aureodatabase>protein 3D view</aureodatabase> | ||
==General== | ==General== | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>protein symbol</aureodatabase> | *<aureodatabase>protein symbol</aureodatabase> | ||
* <aureodatabase>protein description</aureodatabase> | *<aureodatabase>protein description</aureodatabase> | ||
* <aureodatabase>protein length</aureodatabase> | *<aureodatabase>protein length</aureodatabase> | ||
* <aureodatabase>theoretical pI</aureodatabase> | *<aureodatabase>theoretical pI</aureodatabase> | ||
* <aureodatabase>theoretical MW</aureodatabase> | *<aureodatabase>theoretical MW</aureodatabase> | ||
* <aureodatabase>GRAVY</aureodatabase> | *<aureodatabase>GRAVY</aureodatabase> | ||
</protect> | </protect> | ||
| Line 71: | Line 78: | ||
==Function== | ==Function== | ||
* <aureodatabase>protein reaction</aureodatabase> | *<aureodatabase>protein reaction</aureodatabase> | ||
* <aureodatabase>protein TIGRFAM</aureodatabase> | *<aureodatabase>protein TIGRFAM</aureodatabase> | ||
* <aureodatabase>protein TheSeed</aureodatabase> | *<aureodatabase>protein TheSeed</aureodatabase> | ||
* <aureodatabase>protein PFAM</aureodatabase> | *<aureodatabase>protein PFAM</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
==Structure, modifications & | ==Structure, modifications & cofactors== | ||
* <aureodatabase>protein domains</aureodatabase> | *<aureodatabase>protein domains</aureodatabase> | ||
* <aureodatabase>protein modifications</aureodatabase> | *<aureodatabase>protein modifications</aureodatabase> | ||
* <aureodatabase>protein cofactors</aureodatabase> | *<aureodatabase>protein cofactors</aureodatabase> | ||
* <aureodatabase>protein effectors</aureodatabase> | *<aureodatabase>protein effectors</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein regulated operons</aureodatabase> | ||
</protect> | </protect> | ||
| Line 90: | Line 97: | ||
==Localization== | ==Localization== | ||
* <aureodatabase>protein Psortb</aureodatabase> | *<aureodatabase>protein Psortb</aureodatabase> | ||
* <aureodatabase>protein LocateP</aureodatabase> | *<aureodatabase>protein LocateP</aureodatabase> | ||
* <aureodatabase>protein SignalP</aureodatabase> | *<aureodatabase>protein SignalP</aureodatabase> | ||
* <aureodatabase>protein TMHMM</aureodatabase> | *<aureodatabase>protein TMHMM</aureodatabase> | ||
</protect> | </protect> | ||
| Line 99: | Line 106: | ||
==Accession numbers== | ==Accession numbers== | ||
* <aureodatabase>protein GI</aureodatabase> | *<aureodatabase>protein GI</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein RefSeq</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein UniProt</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein STRING</aureodatabase> | ||
</protect> | </protect> | ||
| Line 108: | Line 115: | ||
==Protein sequence== | ==Protein sequence== | ||
* <aureodatabase>protein sequence</aureodatabase> | *<aureodatabase>protein sequence</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
== | ==Experimental data== | ||
* <aureodatabase>protein validated peptides</aureodatabase> | *<aureodatabase>protein validated peptides</aureodatabase> | ||
*<aureodatabase>protein validated localization</aureodatabase> | |||
*<aureodatabase>protein validated quantitative data</aureodatabase> | |||
*<aureodatabase>protein partners</aureodatabase> | |||
</protect> | </protect> | ||
| Line 125: | Line 135: | ||
==Operon== | ==Operon== | ||
* <aureodatabase>operons</aureodatabase> | *<aureodatabase>operons</aureodatabase> | ||
</protect> | </protect> | ||
| Line 131: | Line 141: | ||
==Regulation== | ==Regulation== | ||
*<aureodatabase>regulators</aureodatabase> | |||
* <aureodatabase>regulators</aureodatabase> | |||
</protect> | </protect> | ||
| Line 138: | Line 147: | ||
==Transcription pattern== | ==Transcription pattern== | ||
* <aureodatabase>expression browser</aureodatabase> | *<aureodatabase>expression browser</aureodatabase> | ||
</protect> | </protect> | ||
| Line 144: | Line 153: | ||
==Protein synthesis (provided by Aureolib)== | ==Protein synthesis (provided by Aureolib)== | ||
* <aureodatabase>protein synthesis Aureolib</aureodatabase> | *<aureodatabase>protein synthesis Aureolib</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
== | ==Protein stability== | ||
* <aureodatabase>protein half-life</aureodatabase> | *<aureodatabase>protein half-life</aureodatabase> | ||
</protect> | </protect> | ||
Latest revision as of 23:04, 10 March 2016
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01104
- pan locus tag?: SAUPAN003388000
- symbol: sdhA
- pan gene symbol?: sdhA
- synonym:
- product: succinate dehydrogenase flavoprotein subunit
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01104
- symbol: sdhA
- product: succinate dehydrogenase flavoprotein subunit
- replicon: chromosome
- strand: +
- coordinates: 1065336..1067102
- length: 1767
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920746 NCBI
- RefSeq: YP_499648 NCBI
- BioCyc: G1I0R-1037 BioCyc
- MicrobesOnline: 1289561 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741ATGGCAGAGAAACATCTTATTGTTGTCGGAGGTGGCCTAGCGGGCTTAATGTCAACAATT
AAAGCGGCAGAAAAAGGTGCACATGTAGATTTGTTCTCAGTTGTACCAGTAAAGCGTTCG
CACTCTGTTTGTGCCCAAGGTGGCATTAATGGTGCGGTCAATACTAAAGGGGAAGGCGAT
TCTCCTTGGATTCACTTTGATGATACAGTGTATGGTGGCGATTTCCTTGCAAACCAACCA
CCTGTTAAAGCGATGACAGAGGCAGCACCTAAAATTATTCATTTATTAGACCGTATGGGC
GTAATGTTCAATAGAACAAATGAAGGTCTATTAGATTTTAGACGTTTCGGTGGTACATTA
CATCACAGAACAGCATATGCAGGGGCAACAACTGGACAACAATTATTATATGCATTGGAT
GAACAAGTTCGTGCATATGAAGTAGATGGATTAGTTACGAAGTATGAAGGATGGGAATTC
CTTGGCATAGTTAAAGGTGACGATGATAGTGCAAGAGGTATCGTTGCACAAAATATGACA
ACTGCTGAGATTGAAACATTTGGTTCAGATGCAGTTATTATGGCAACGGGTGGCCCTGGT
ATTATTTTCGGTAAAACAACAAACTCAATGATTAATACAGGATCAGCGGCTTCCATTGTT
TACCAACAAGGCGCTATTTATGCTAATGGTGAGTTCATTCAAATTCATCCTACTGCAATC
CCTGGTGATGATAAACTGCGACTAATGAGTGAATCAGCACGTGGTGAAGGTGGACGAATT
TGGACATATAAAGATGGTAAGCCTTGGTACTTCTTAGAAGAGAAATATCCTGATTATGGT
AACTTAGTACCTCGTGATATCGCAACGCGTGAAATTTTCGATGTATGTATTAACCAAAAA
TTAGGTATAAATGGCGAAAACATGGTATATCTTGATTTGTCACATAAAGATCCACATGAG
TTAGATGTAAAACTAGGTGGTATCATTGAGATTTATGAAAAATTCACTGGTGATGACCCA
CGCAAAGTACCAATGAAGATTTTCCCAGCTGTTCACTATTCAATGGGTGGTCTATATGTA
GATTATGATCAAATGACAAATATTAAAGGGTTATTTGCAGCTGGAGAATGTGACTTCTCT
CAACATGGTGGTAACCGCTTAGGTGCCAATTCATTGTTATCAGCGATTTATGGTGGTACA
GTAGCAGGTCCAAACGCGATTGATTATATTTCAAATATTGATCGATCATATACTGATATG
GACGAAAGTATTTTTGAAAAGCGTAAAGCTGAAGAGCAAGAACGTTTTGATAAATTATTA
GCTATGCGCGGTACAGAAAATGCATATAAATTACACCGTGAACTTGGTGAAATTATGACA
GCAAATGTAACTGTTGTTCGTGAAAATGAAAAACTGTTAGAAACAGATAAAAAGATTGTT
GAATTGATGAAACGTTATGAAGATATTGATATGGAAGATACTCAAACTTGGAGTAACCAA
GCGGTATTCTTTACCCGTCAACTATGGAACATGTTAGTACTTGCACGTGTTATTACGATT
GGTGCATATAACCGTAACGAATCACGCGGTGCCCATTATAAACCAGAATTCCCAGAGCGT
AATGATGAAGAGTGGTTAAAAACGACAATGGCCTCATTCCAAGGCGCATTTGAAAAACCG
CAGTTTACTTATGATGACGTCGATGTGAGTTTAATACCACCTCGTAAACGTGATTACACA
AGTAAGTCTAAAGGGGGTAAAAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1767
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01104
- symbol: SdhA
- description: succinate dehydrogenase flavoprotein subunit
- length: 588
- theoretical pI: 5.3235
- theoretical MW: 65502.6
- GRAVY: -0.398129
⊟Function[edit | edit source]
- reaction: EC 1.3.5.1? ExPASySuccinate dehydrogenase (quinone) Succinate + a quinone = fumarate + a quinolEC 1.3.99.1? ExPASyDeleted entry
- TIGRFAM: Energy metabolism TCA cycle succinate dehydrogenase or fumarate reductase, flavoprotein subunit (TIGR01811; EC 1.3.99.1; HMM-score: 970.9)and 25 moreEnergy metabolism TCA cycle succinate dehydrogenase or fumarate reductase, flavoprotein subunit (TIGR01812; HMM-score: 475.7)Energy metabolism Anaerobic succinate dehydrogenase or fumarate reductase, flavoprotein subunit (TIGR01812; HMM-score: 475.7)Energy metabolism Aerobic succinate dehydrogenase or fumarate reductase, flavoprotein subunit (TIGR01812; HMM-score: 475.7)Energy metabolism TCA cycle succinate dehydrogenase, flavoprotein subunit (TIGR01816; HMM-score: 364.7)fumarate reductase (quinol), flavoprotein subunit (TIGR01176; EC 1.3.5.4; HMM-score: 333.1)Biosynthesis of cofactors, prosthetic groups, and carriers Pyridine nucleotides L-aspartate oxidase (TIGR00551; EC 1.4.3.16; HMM-score: 274.9)Energy metabolism Electron transport flavocytochrome c (TIGR01813; HMM-score: 119)Unknown function Enzymes of unknown specificity flavoprotein, HI0933 family (TIGR00275; HMM-score: 19.5)Energy metabolism Electron transport thioredoxin-disulfide reductase (TIGR01292; EC 1.8.1.9; HMM-score: 17.2)Central intermediary metabolism Sulfur metabolism adenylylsulfate reductase, alpha subunit (TIGR02061; EC 1.8.99.2; HMM-score: 15.5)squalene-associated FAD-dependent desaturase (TIGR03467; HMM-score: 14.3)Cellular processes Detoxification alkyl hydroperoxide reductase subunit F (TIGR03140; EC 1.8.1.-; HMM-score: 14.2)Cellular processes Adaptations to atypical conditions alkyl hydroperoxide reductase subunit F (TIGR03140; EC 1.8.1.-; HMM-score: 14.2)Biosynthesis of cofactors, prosthetic groups, and carriers Heme, porphyrin, and cobalamin precorrin 3B synthase CobZ (TIGR02485; HMM-score: 14.1)Biosynthesis of cofactors, prosthetic groups, and carriers Chlorophyll and bacteriochlorphyll geranylgeranyl reductase family (TIGR02032; EC 1.3.1.-; HMM-score: 13.7)Cellular processes Detoxification mercury(II) reductase (TIGR02053; EC 1.16.1.1; HMM-score: 13.5)Protein synthesis tRNA and rRNA base modification tRNA U-34 5-methylaminomethyl-2-thiouridine biosynthesis protein MnmC, C-terminal domain (TIGR03197; HMM-score: 12.9)Biosynthesis of cofactors, prosthetic groups, and carriers Thiamine glycine oxidase ThiO (TIGR02352; EC 1.4.3.19; HMM-score: 12.7)Energy metabolism Anaerobic glycerol-3-phosphate dehydrogenase, anaerobic, B subunit (TIGR03378; EC 1.1.5.3; HMM-score: 12.5)FAD dependent oxidoreductase TIGR03364 (TIGR03364; HMM-score: 11.7)Energy metabolism Amino acids and amines sarcosine oxidase, alpha subunit family (TIGR01372; HMM-score: 11.3)mycofactocin system FadH/OYE family oxidoreductase 2 (TIGR03997; EC 1.-.-.-; HMM-score: 11.1)thioredoxin and glutathione reductase (TIGR01438; EC 1.6.4.-; HMM-score: 10.9)Amino acid biosynthesis Glutamate family glutamate synthase (NADPH), homotetrameric (TIGR01316; EC 1.4.1.13; HMM-score: 10.2)dihydrolipoyl dehydrogenase (TIGR01350; EC 1.8.1.4; HMM-score: 9.6)
- TheSEED :
- Succinate dehydrogenase flavoprotein subunit (EC 1.3.99.1)
Carbohydrates Central carbohydrate metabolism TCA Cycle Succinate dehydrogenase flavoprotein subunit (EC 1.3.99.1)and 2 more - PFAM: NADP_Rossmann (CL0063) FAD_binding_2; FAD binding domain (PF00890; HMM-score: 283.8)and 8 moreno clan defined Succ_DH_flav_C; Fumarate reductase flavoprotein C-term (PF02910; HMM-score: 141.3)NADP_Rossmann (CL0063) Pyr_redox_2; Pyridine nucleotide-disulphide oxidoreductase (PF07992; HMM-score: 24)GIDA; Glucose inhibited division protein A (PF01134; HMM-score: 19.3)HI0933_like; HI0933-like protein (PF03486; HMM-score: 18.5)FAD_oxidored; FAD dependent oxidoreductase (PF12831; HMM-score: 14.9)AlaDh_PNT_C; Alanine dehydrogenase/PNT, C-terminal domain (PF01262; HMM-score: 14.3)Pyr_redox; Pyridine nucleotide-disulphide oxidoreductase (PF00070; HMM-score: 14)NAD_binding_8; NAD(P)-binding Rossmann-like domain (PF13450; HMM-score: 13)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.89
- Cytoplasmic Membrane Score: 0.09
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- LocateP: N-terminally anchored (No CS)
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.5
- Signal peptide possibility: 0
- N-terminally Anchored Score: 9
- Predicted Cleavage Site: No CleavageSite
- SignalP: Signal peptide SP(Sec/SPI) length 22 aa
- SP(Sec/SPI): 0.583697
- TAT(Tat/SPI): 0.035658
- LIPO(Sec/SPII): 0.011556
- Cleavage Site: CS pos: 22-23. IKA-AE. Pr: 0.4744
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MAEKHLIVVGGGLAGLMSTIKAAEKGAHVDLFSVVPVKRSHSVCAQGGINGAVNTKGEGDSPWIHFDDTVYGGDFLANQPPVKAMTEAAPKIIHLLDRMGVMFNRTNEGLLDFRRFGGTLHHRTAYAGATTGQQLLYALDEQVRAYEVDGLVTKYEGWEFLGIVKGDDDSARGIVAQNMTTAEIETFGSDAVIMATGGPGIIFGKTTNSMINTGSAASIVYQQGAIYANGEFIQIHPTAIPGDDKLRLMSESARGEGGRIWTYKDGKPWYFLEEKYPDYGNLVPRDIATREIFDVCINQKLGINGENMVYLDLSHKDPHELDVKLGGIIEIYEKFTGDDPRKVPMKIFPAVHYSMGGLYVDYDQMTNIKGLFAAGECDFSQHGGNRLGANSLLSAIYGGTVAGPNAIDYISNIDRSYTDMDESIFEKRKAEEQERFDKLLAMRGTENAYKLHRELGEIMTANVTVVRENEKLLETDKKIVELMKRYEDIDMEDTQTWSNQAVFFTRQLWNMLVLARVITIGAYNRNESRGAHYKPEFPERNDEEWLKTTMASFQGAFEKPQFTYDDVDVSLIPPRKRDYTSKSKGGKK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_01103 > sdhA > sdhBpredicted SigA promoter [3] : S455 > SAOUHSC_01103 > sdhA > sdhB > S456 > SAOUHSC_01106 > SAOUHSC_01107 > SAOUHSC_01108predicted SigB promoter [3] : S455 > SAOUHSC_01103 > sdhA > sdhB > S456 > SAOUHSC_01106 > SAOUHSC_01107 > SAOUHSC_01108
⊟Regulation[edit | edit source]
- regulator: CcpA* regulon
CcpA* (TF) important in Carbon catabolism; RegPrecise transcription unit transferred from N315 data RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
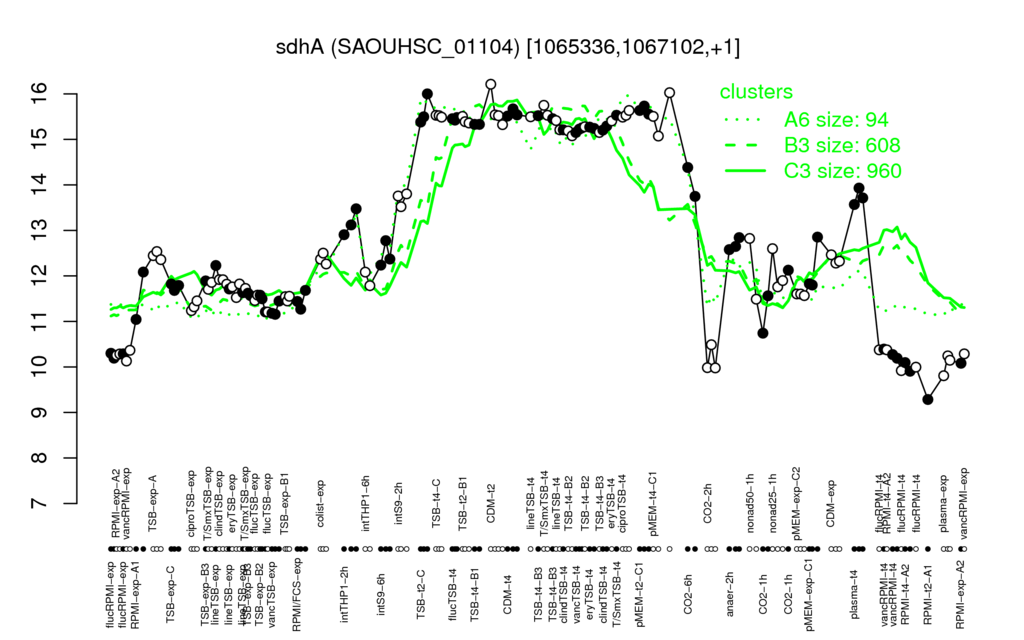 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You are kindly invited to share additional interesting facts.
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Rosmarie Gaupp, Steffen Schlag, Manuel Liebeke, Michael Lalk, Friedrich Götz
Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms.
J Bacteriol: 2010, 192(9);2385-94
[PubMed:20207757] [WorldCat.org] [DOI] (I p)
